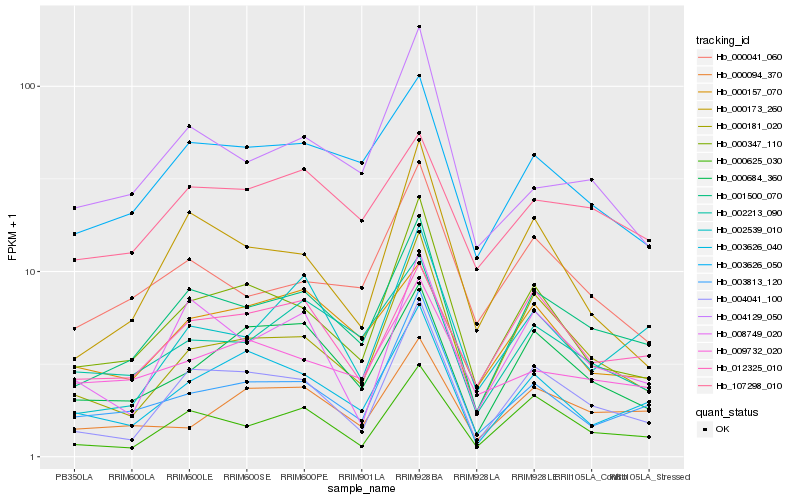
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001500_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003626_050 |
0.1080236487 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 3 |
Hb_004041_100 |
0.1179766642 |
- |
- |
- |
| 4 |
Hb_002213_090 |
0.1181342529 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 5 |
Hb_000625_030 |
0.1211820137 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 6 |
Hb_000347_110 |
0.1255462366 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 7 |
Hb_012325_010 |
0.1277125144 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 8 |
Hb_000684_360 |
0.1304575574 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 9 |
Hb_107298_010 |
0.1323649058 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 10 |
Hb_000041_060 |
0.1334304869 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 11 |
Hb_003626_040 |
0.1337038437 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000181_020 |
0.1351748997 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_008749_020 |
0.1379411307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003813_120 |
0.1421713387 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_002539_010 |
0.1427830503 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 16 |
Hb_000094_370 |
0.142814941 |
- |
- |
- |
| 17 |
Hb_000173_260 |
0.1429126094 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 18 |
Hb_000157_070 |
0.1430480445 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 19 |
Hb_004129_050 |
0.1436326645 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 5 [Jatropha curcas] |
| 20 |
Hb_009732_020 |
0.1478383522 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |