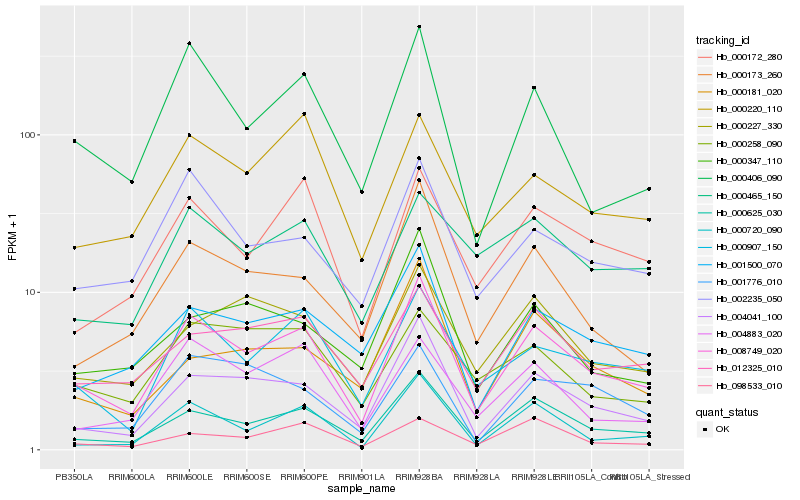
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008749_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000625_030 |
0.083977629 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 3 |
Hb_004041_100 |
0.1102978437 |
- |
- |
- |
| 4 |
Hb_000907_150 |
0.1211552338 |
- |
- |
hypothetical protein L484_004295 [Morus notabilis] |
| 5 |
Hb_000406_090 |
0.1302450993 |
- |
- |
unknown [Populus trichocarpa] |
| 6 |
Hb_000720_090 |
0.1327572923 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 7 |
Hb_001500_070 |
0.1379411307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002235_050 |
0.1397294275 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000173_260 |
0.1417205852 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 10 |
Hb_000172_280 |
0.1434476167 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 11 |
Hb_012325_010 |
0.1462396695 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 12 |
Hb_004883_020 |
0.1531042595 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 13 |
Hb_000227_330 |
0.1548276912 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 14 |
Hb_000220_110 |
0.1551242021 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 15 |
Hb_098533_010 |
0.1561624771 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 16 |
Hb_000181_020 |
0.162430198 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000347_110 |
0.1625062766 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 18 |
Hb_000258_090 |
0.1632022376 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000465_150 |
0.1654405644 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 20 |
Hb_001776_010 |
0.1656362329 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |