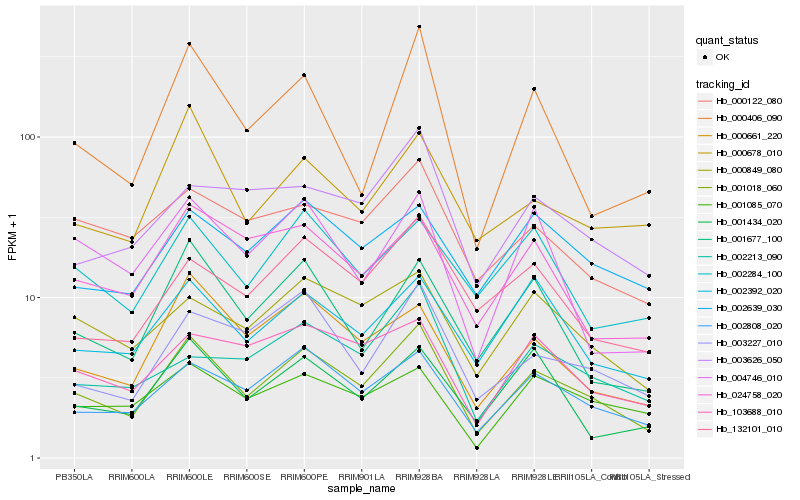
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001018_060 |
0.0 |
- |
- |
hypothetical protein B456_001G028000 [Gossypium raimondii] |
| 2 |
Hb_002808_020 |
0.1073499097 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000661_220 |
0.1304025147 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 4 |
Hb_000849_080 |
0.1322773042 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003227_010 |
0.1335167167 |
- |
- |
PREDICTED: dynamin-related protein 1E-like [Jatropha curcas] |
| 6 |
Hb_002392_020 |
0.135077824 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 7 |
Hb_001677_100 |
0.136247399 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 8 |
Hb_103688_010 |
0.1402088199 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 9 |
Hb_132101_010 |
0.1410320306 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 10 |
Hb_000678_010 |
0.1416730623 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000122_080 |
0.1420389136 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 12 |
Hb_000406_090 |
0.1422291549 |
- |
- |
unknown [Populus trichocarpa] |
| 13 |
Hb_002639_030 |
0.144023419 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 14 |
Hb_024758_020 |
0.1440683451 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 15 |
Hb_001085_070 |
0.1448730505 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 16 |
Hb_002284_100 |
0.1455771806 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 17 |
Hb_003626_050 |
0.1477276396 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 18 |
Hb_001434_020 |
0.148368685 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 19 |
Hb_002213_090 |
0.1492530545 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 20 |
Hb_004746_010 |
0.1516766199 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |