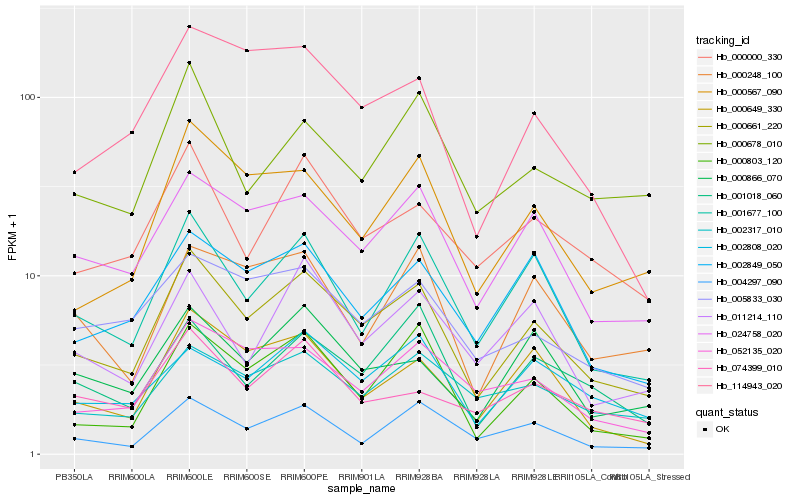
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_220 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 2 |
Hb_024758_020 |
0.1003267767 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 3 |
Hb_001677_100 |
0.1122277357 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 4 |
Hb_000567_090 |
0.117144315 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000000_330 |
0.1243930002 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 6 |
Hb_000803_120 |
0.1246976046 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002849_050 |
0.1267146916 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 8 |
Hb_002317_010 |
0.1271928587 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 9 |
Hb_000649_330 |
0.1276395312 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 10 |
Hb_052135_020 |
0.128987329 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 11 |
Hb_002808_020 |
0.1290782147 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001018_060 |
0.1304025147 |
- |
- |
hypothetical protein B456_001G028000 [Gossypium raimondii] |
| 13 |
Hb_000866_070 |
0.1304903049 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 14 |
Hb_005833_030 |
0.1315131432 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 15 |
Hb_114943_020 |
0.134820489 |
- |
- |
PREDICTED: NADPH--cytochrome P450 reductase 2 [Jatropha curcas] |
| 16 |
Hb_004297_090 |
0.135082005 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 17 |
Hb_000678_010 |
0.1363997886 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_074399_010 |
0.1366808865 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 19 |
Hb_000248_100 |
0.1392080369 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 20 |
Hb_011214_110 |
0.1397602918 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |