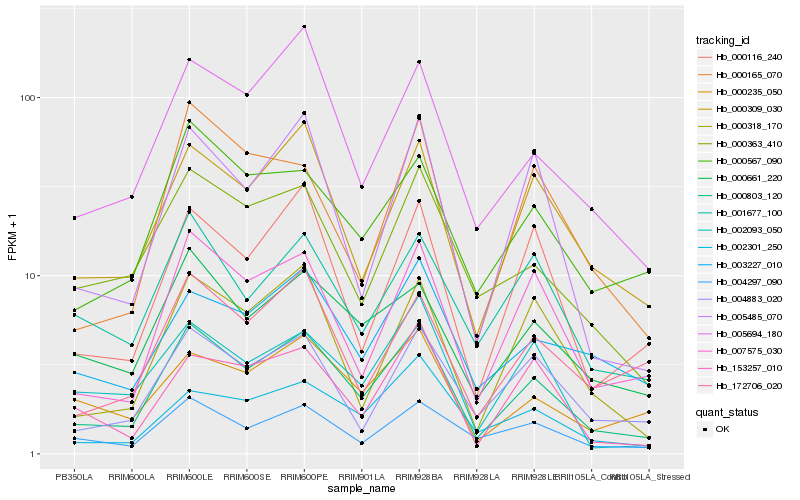
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_120 |
0.0 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_007575_030 |
0.1127366007 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 3 |
Hb_000309_030 |
0.1131004103 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_005694_180 |
0.1232876669 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 5 |
Hb_000661_220 |
0.1246976046 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 6 |
Hb_004297_090 |
0.1261920276 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 7 |
Hb_000567_090 |
0.1314249217 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_005485_070 |
0.1314967404 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000363_410 |
0.1319758085 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 10 |
Hb_002301_250 |
0.1323576059 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a [Jatropha curcas] |
| 11 |
Hb_004883_020 |
0.1335949188 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 12 |
Hb_000116_240 |
0.1346302164 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 13 |
Hb_000318_170 |
0.1353198707 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 14 |
Hb_001677_100 |
0.1402762423 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 15 |
Hb_000235_050 |
0.1404510245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003227_010 |
0.1424034427 |
- |
- |
PREDICTED: dynamin-related protein 1E-like [Jatropha curcas] |
| 17 |
Hb_153257_010 |
0.1439176772 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 18 |
Hb_172706_020 |
0.1446580591 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 19 |
Hb_002093_050 |
0.1450012502 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 20 |
Hb_000165_070 |
0.1451403894 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |