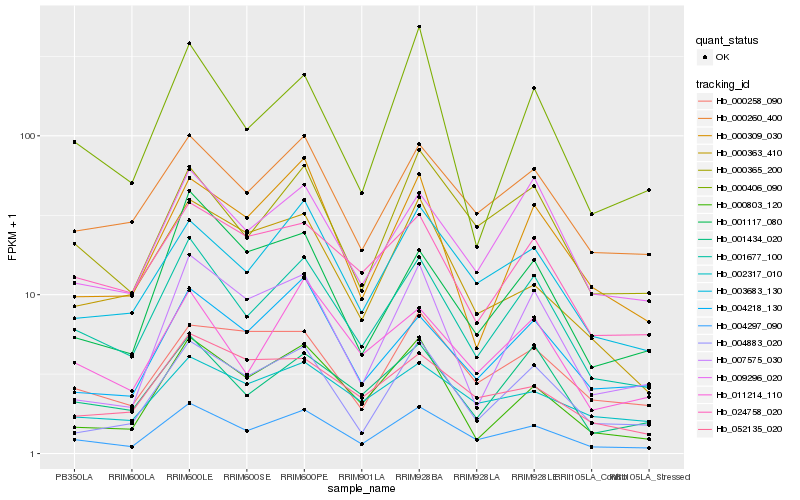
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004297_090 |
0.0 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 2 |
Hb_001677_100 |
0.0703826176 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 3 |
Hb_000365_200 |
0.0906940245 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 4 |
Hb_003683_130 |
0.0929643397 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000260_400 |
0.103361658 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 6 |
Hb_004883_020 |
0.1054894827 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 7 |
Hb_009296_020 |
0.1216102399 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_004218_130 |
0.121867308 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 9 |
Hb_000258_090 |
0.1240741884 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000363_410 |
0.1251206527 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 11 |
Hb_000803_120 |
0.1261920276 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000309_030 |
0.1289510749 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_011214_110 |
0.1312786657 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 14 |
Hb_002317_010 |
0.1319278182 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_001434_020 |
0.1323800233 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 16 |
Hb_000406_090 |
0.1340257179 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_007575_030 |
0.1341673818 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 18 |
Hb_001117_080 |
0.1344171254 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_052135_020 |
0.1344593004 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 20 |
Hb_024758_020 |
0.1350351169 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |