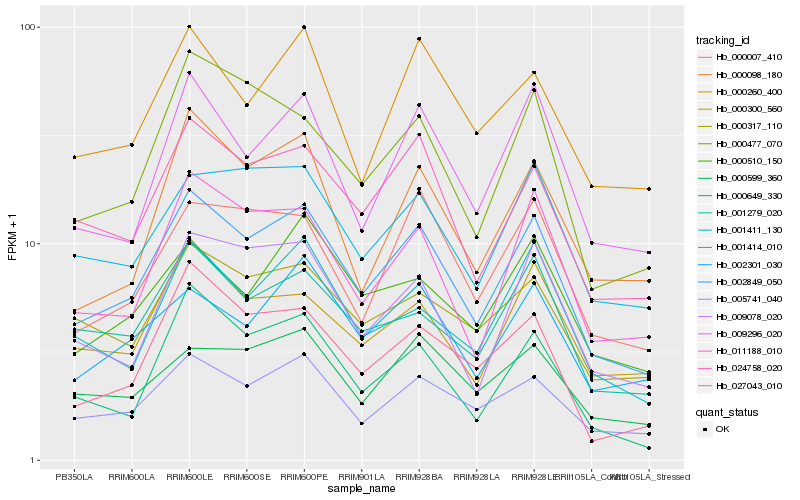
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002849_050 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 2 |
Hb_005741_040 |
0.0784553045 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 3 |
Hb_024758_020 |
0.0816562349 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 4 |
Hb_011188_010 |
0.0896587344 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009296_020 |
0.0942038408 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_001279_020 |
0.0964973005 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 7 |
Hb_001414_010 |
0.0971665738 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 8 |
Hb_009078_020 |
0.0987658881 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 9 |
Hb_000317_110 |
0.103186568 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 10 |
Hb_000300_560 |
0.1037108263 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000477_070 |
0.1040166986 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 12 |
Hb_000007_410 |
0.1042313991 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 13 |
Hb_002301_030 |
0.1044676614 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 14 |
Hb_000649_330 |
0.104881287 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 15 |
Hb_027043_010 |
0.1052626731 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase GDPD4 [Jatropha curcas] |
| 16 |
Hb_001411_130 |
0.1054875222 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000599_360 |
0.1057091687 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 18 |
Hb_000260_400 |
0.1060944725 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 19 |
Hb_000098_180 |
0.107315608 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000510_150 |
0.1080617095 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |