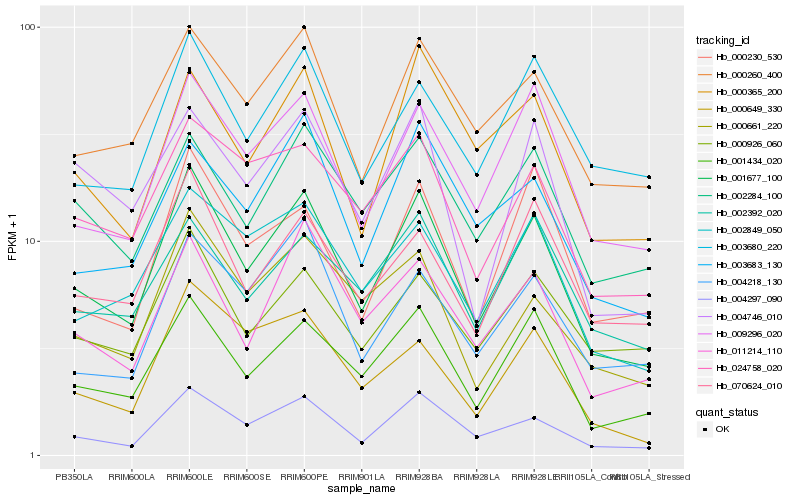
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001677_100 |
0.0 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 2 |
Hb_004297_090 |
0.0703826176 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 3 |
Hb_001434_020 |
0.0808686949 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 4 |
Hb_009296_020 |
0.0937687422 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000260_400 |
0.1020621502 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 6 |
Hb_011214_110 |
0.105997934 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 7 |
Hb_002849_050 |
0.1096695643 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 8 |
Hb_024758_020 |
0.1105639255 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 9 |
Hb_070624_010 |
0.1115972402 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 10 |
Hb_000661_220 |
0.1122277357 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 11 |
Hb_000365_200 |
0.1145047241 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 12 |
Hb_002284_100 |
0.1146552533 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 13 |
Hb_000230_530 |
0.1153688259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003683_130 |
0.117237626 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000926_060 |
0.117467423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004746_010 |
0.1183285313 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 17 |
Hb_003680_220 |
0.1211401087 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 18 |
Hb_002392_020 |
0.1245923715 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 19 |
Hb_000649_330 |
0.1250453381 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 20 |
Hb_004218_130 |
0.1256895509 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |