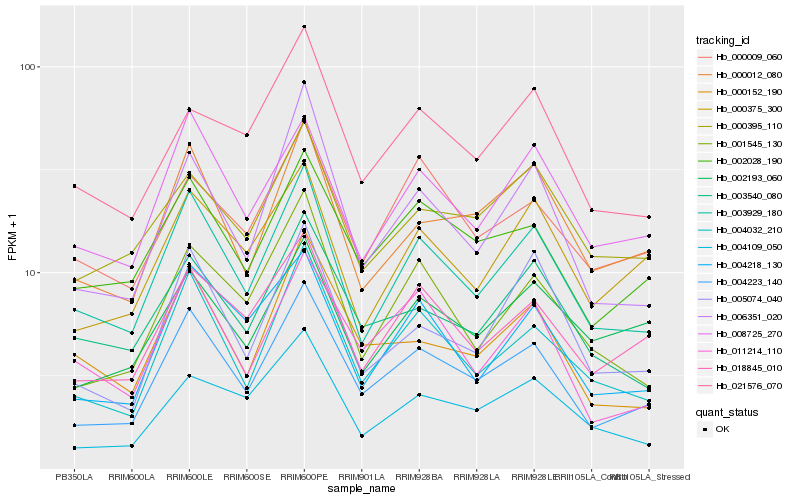
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_140 |
0.0 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 2 |
Hb_003929_180 |
0.0791734715 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 3 |
Hb_002028_190 |
0.0827105964 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7-like [Jatropha curcas] |
| 4 |
Hb_004032_210 |
0.0828092508 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_018845_010 |
0.0920497551 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 6 |
Hb_000012_080 |
0.0980996678 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 7 |
Hb_006351_020 |
0.100060329 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 8 |
Hb_004218_130 |
0.1018392004 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 9 |
Hb_008725_270 |
0.1022362047 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 10 |
Hb_011214_110 |
0.104969794 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 11 |
Hb_000152_190 |
0.1084183858 |
- |
- |
PREDICTED: K(+) efflux antiporter 5 [Jatropha curcas] |
| 12 |
Hb_001545_130 |
0.1101348432 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 13 |
Hb_004109_050 |
0.1111356671 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 14 |
Hb_003540_080 |
0.1131284731 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_005074_040 |
0.1141961566 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000375_300 |
0.1145252005 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 17 |
Hb_000395_110 |
0.1156033306 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 18 |
Hb_002193_060 |
0.1168114418 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 19 |
Hb_000009_060 |
0.117111262 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 20 |
Hb_021576_070 |
0.1181663945 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |