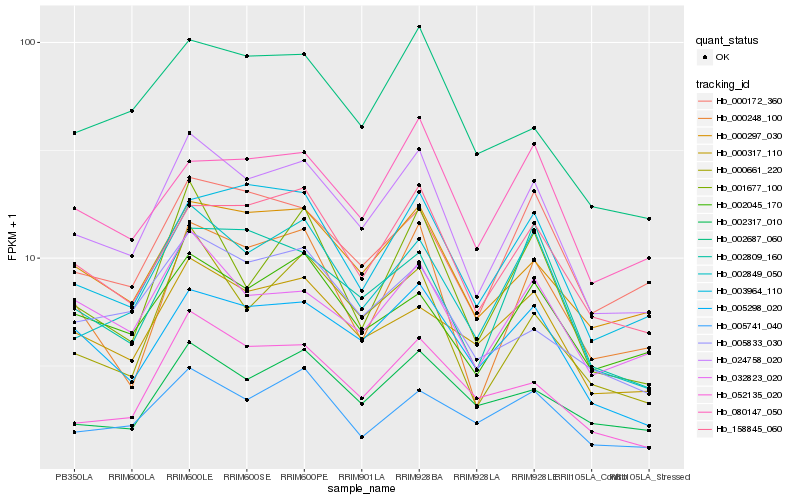
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024758_020 |
0.0 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 2 |
Hb_002849_050 |
0.0816562349 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 3 |
Hb_158845_060 |
0.0834087341 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005298_020 |
0.0865151923 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 2 [Jatropha curcas] |
| 5 |
Hb_000297_030 |
0.0891457282 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_032823_020 |
0.0960223281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003964_110 |
0.0993448819 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000661_220 |
0.1003267767 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 9 |
Hb_000317_110 |
0.100560313 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 10 |
Hb_000172_360 |
0.1033049738 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 11 |
Hb_002045_170 |
0.1063079987 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 12 |
Hb_002809_160 |
0.1070600779 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 13 |
Hb_002687_060 |
0.1082739397 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 14 |
Hb_005833_030 |
0.1082992716 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 15 |
Hb_002317_010 |
0.1088185836 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_080147_050 |
0.1088712225 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 17 |
Hb_000248_100 |
0.1091334979 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 18 |
Hb_005741_040 |
0.1093021023 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 19 |
Hb_052135_020 |
0.10982991 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 20 |
Hb_001677_100 |
0.1105639255 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |