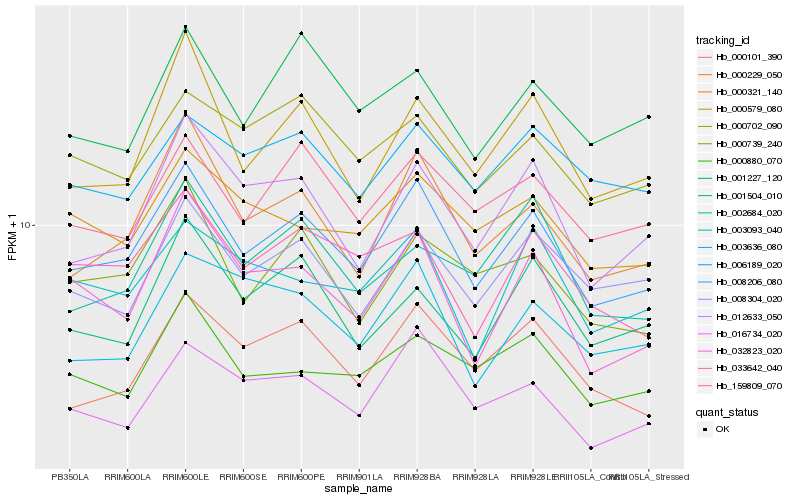
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008206_080 |
0.0 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 17 [Jatropha curcas] |
| 2 |
Hb_000229_050 |
0.0654336356 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 3 |
Hb_000579_080 |
0.0743516883 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 4 |
Hb_012633_050 |
0.0772141814 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_159809_070 |
0.0777768037 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 6 |
Hb_008304_020 |
0.0799753469 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 7 |
Hb_000101_390 |
0.0827883212 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 8 |
Hb_033642_040 |
0.0829271078 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000739_240 |
0.0833567419 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_016734_020 |
0.0845544578 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_032823_020 |
0.0853418823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000880_070 |
0.0871177004 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 13 |
Hb_000702_090 |
0.0892823784 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 14 |
Hb_000321_140 |
0.0894328814 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 15 |
Hb_003093_040 |
0.0905575871 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_006189_020 |
0.0909439791 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 17 |
Hb_001227_120 |
0.0933605173 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 18 |
Hb_001504_010 |
0.094403981 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 19 |
Hb_003636_080 |
0.0957666503 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 20 |
Hb_002684_020 |
0.096470521 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |