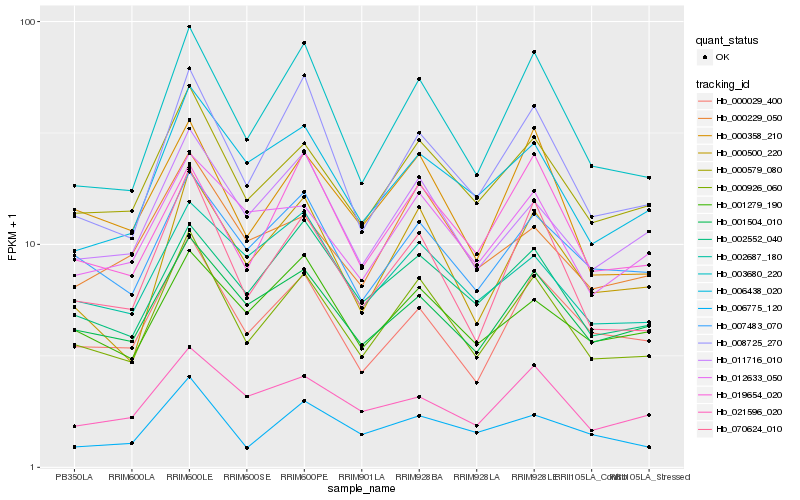
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000926_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000579_080 |
0.0596758171 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 3 |
Hb_070624_010 |
0.0639389755 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 4 |
Hb_003680_220 |
0.0679754144 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 5 |
Hb_000500_220 |
0.0797158524 |
- |
- |
PREDICTED: intersectin-1 [Jatropha curcas] |
| 6 |
Hb_008725_270 |
0.0803207862 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 7 |
Hb_000358_210 |
0.0830632531 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 8 |
Hb_000029_400 |
0.0841076563 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011716_010 |
0.0848972349 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 10 |
Hb_007483_070 |
0.0850661348 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 11 |
Hb_001504_010 |
0.0862748215 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 12 |
Hb_002552_040 |
0.0938463097 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 13 |
Hb_006438_020 |
0.0942711163 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 14 |
Hb_006775_120 |
0.0973259004 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 15 |
Hb_019654_020 |
0.0985180716 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 16 |
Hb_000229_050 |
0.1000666487 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 17 |
Hb_012633_050 |
0.1003220852 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_021596_020 |
0.1005736382 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 19 |
Hb_001279_190 |
0.1007761903 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002687_180 |
0.1008455454 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |