| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
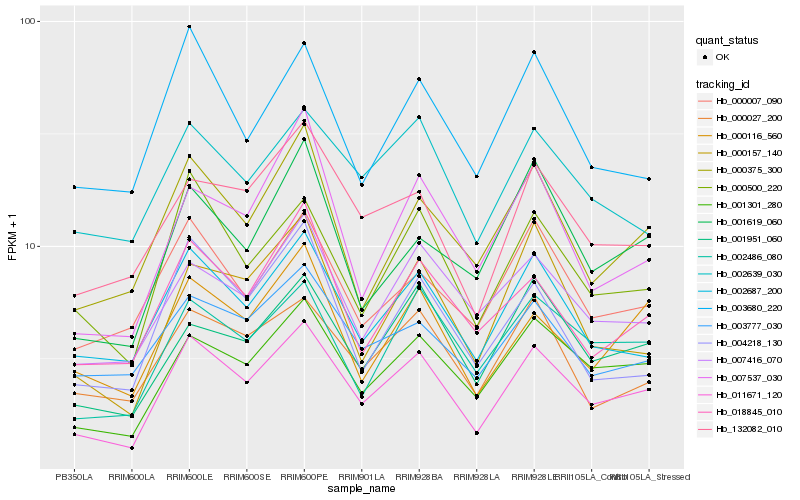
Hb_011671_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_26275 [Jatropha curcas] |
| 2 |
Hb_001301_280 |
0.0808125765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000500_220 |
0.0925180499 |
- |
- |
PREDICTED: intersectin-1 [Jatropha curcas] |
| 4 |
Hb_002687_200 |
0.0999609556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002486_080 |
0.1010267029 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 6 |
Hb_001951_060 |
0.105867152 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 7 |
Hb_000375_300 |
0.1084438251 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 8 |
Hb_000116_560 |
0.1116315426 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 9 |
Hb_018845_010 |
0.1116920781 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 10 |
Hb_132082_010 |
0.1118037771 |
- |
- |
hypothetical protein VITISV_030895 [Vitis vinifera] |
| 11 |
Hb_000157_140 |
0.1140471881 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 12 |
Hb_000027_200 |
0.1147725922 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 13 |
Hb_007416_070 |
0.1149515025 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 14 |
Hb_000007_090 |
0.1159294253 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 15 |
Hb_003680_220 |
0.1175811386 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 16 |
Hb_001619_060 |
0.1185514074 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007537_030 |
0.120008335 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002639_030 |
0.1208550757 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 19 |
Hb_004218_130 |
0.1228468084 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 20 |
Hb_003777_030 |
0.1231040704 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |