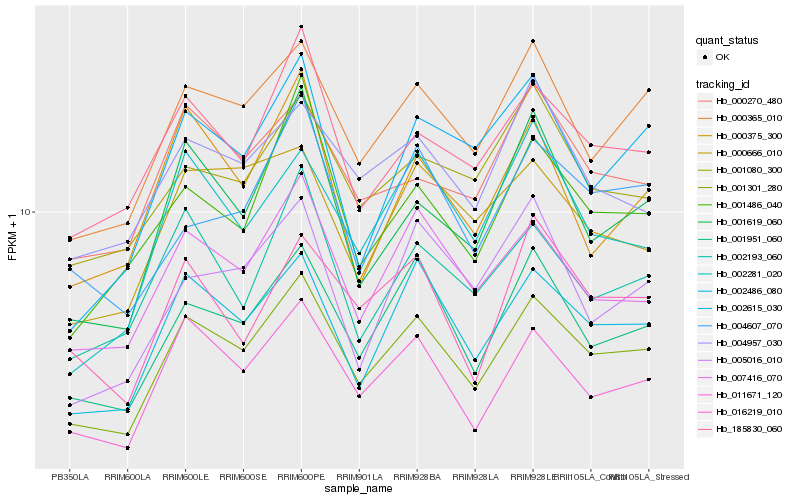
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001301_280 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002486_080 |
0.0686082227 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 3 |
Hb_001951_060 |
0.0737087407 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 4 |
Hb_000365_010 |
0.0802966852 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 5 |
Hb_011671_120 |
0.0808125765 |
- |
- |
hypothetical protein JCGZ_26275 [Jatropha curcas] |
| 6 |
Hb_007416_070 |
0.0885374848 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 7 |
Hb_001619_060 |
0.0962294284 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 8 |
Hb_185830_060 |
0.1003992038 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 9 |
Hb_002281_020 |
0.1014807273 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 10 |
Hb_002615_030 |
0.1055704682 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 11 |
Hb_002193_060 |
0.1059977751 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 12 |
Hb_001486_040 |
0.1080643807 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004607_070 |
0.108529712 |
- |
- |
integral membrane protein, putative [Ricinus communis] |
| 14 |
Hb_016219_010 |
0.1107472535 |
- |
- |
hypothetical protein B456_001G213800 [Gossypium raimondii] |
| 15 |
Hb_000270_480 |
0.1119466957 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 16 |
Hb_000666_010 |
0.11202793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000375_300 |
0.1140424862 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 18 |
Hb_004957_030 |
0.1148828858 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 19 |
Hb_001080_300 |
0.1152634734 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 20 |
Hb_005016_010 |
0.1155604739 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |