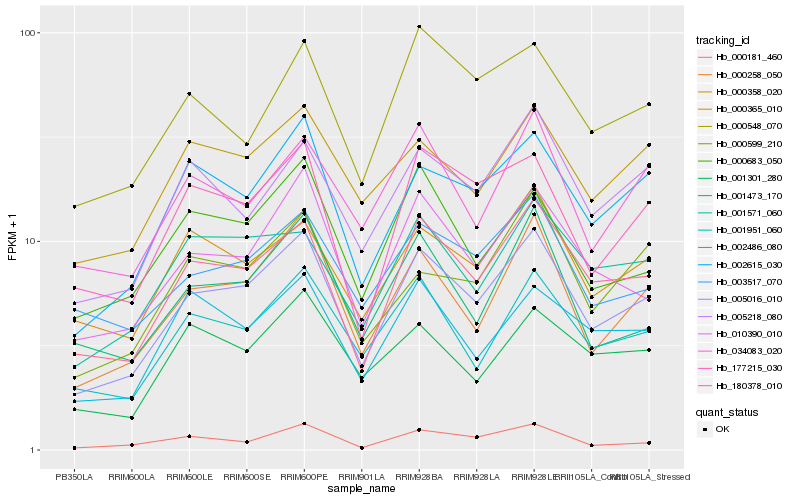
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005016_010 |
0.0 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 2 |
Hb_180378_010 |
0.0682542443 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 3 |
Hb_000258_050 |
0.0763784457 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002615_030 |
0.0844538676 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 5 |
Hb_010390_010 |
0.0911296094 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 6 |
Hb_001951_060 |
0.0958897709 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 7 |
Hb_177215_030 |
0.0996090999 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_005218_080 |
0.1031606872 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000365_010 |
0.1056223505 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 10 |
Hb_000358_020 |
0.1069374718 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 11 |
Hb_003517_070 |
0.1073100248 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 12 |
Hb_001571_060 |
0.1079562514 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 13 |
Hb_000181_460 |
0.1080645311 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000599_210 |
0.1094283038 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000548_070 |
0.1095885434 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 16 |
Hb_001473_170 |
0.109918691 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 17 |
Hb_000683_050 |
0.1113111226 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 18 |
Hb_002486_080 |
0.1115108563 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 19 |
Hb_034083_020 |
0.1124181237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001301_280 |
0.1155604739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |