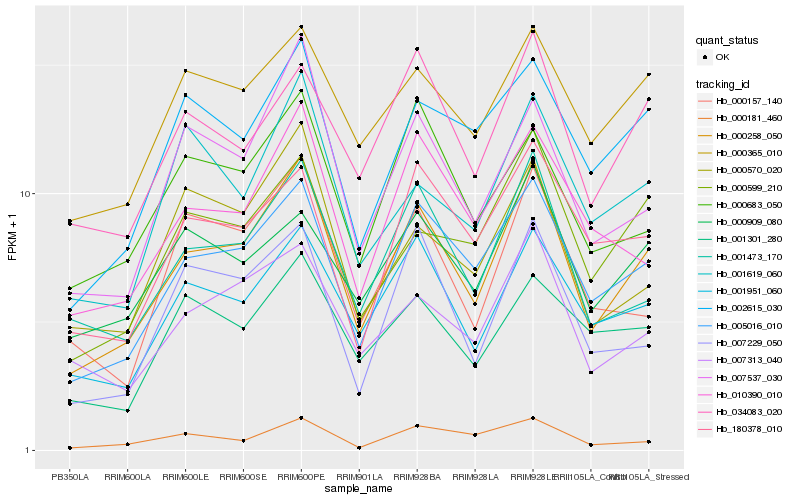
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_050 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005016_010 |
0.0763784457 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 3 |
Hb_001473_170 |
0.0929433569 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 4 |
Hb_001951_060 |
0.1009148332 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 5 |
Hb_007313_040 |
0.1088283575 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 6 |
Hb_007537_030 |
0.1089892595 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_034083_020 |
0.1140545416 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000599_210 |
0.1146422944 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 9 |
Hb_010390_010 |
0.1170919619 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 10 |
Hb_002615_030 |
0.1174546173 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 11 |
Hb_000365_010 |
0.1190042619 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 12 |
Hb_007229_050 |
0.1207383876 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000157_140 |
0.1207667719 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_000683_050 |
0.1226574919 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 15 |
Hb_000909_080 |
0.1235004149 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 16 |
Hb_180378_010 |
0.1238693296 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 17 |
Hb_000570_020 |
0.1245268624 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 18 |
Hb_000181_460 |
0.1256877576 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001619_060 |
0.1274932797 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001301_280 |
0.1293674597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |