| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
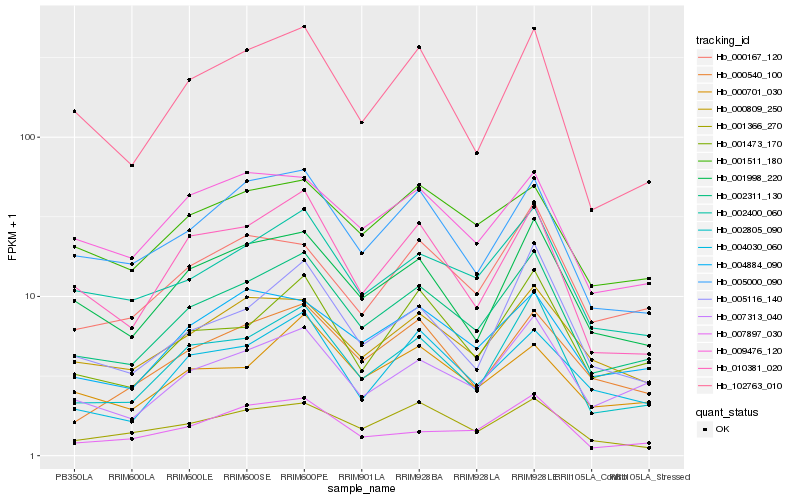
Hb_002311_130 |
0.0 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002805_090 |
0.0677302287 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 3 |
Hb_002400_060 |
0.0765269799 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 4 |
Hb_007313_040 |
0.0792627971 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 5 |
Hb_001998_220 |
0.0865950795 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001473_170 |
0.091400481 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 7 |
Hb_005000_090 |
0.0918582158 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 8 |
Hb_010381_020 |
0.0963541445 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 9 |
Hb_102763_010 |
0.0964555058 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 10 |
Hb_000809_250 |
0.096795032 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 11 |
Hb_004884_090 |
0.0985908276 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 12 |
Hb_001366_270 |
0.1004637481 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 13 |
Hb_005116_140 |
0.1017633447 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 14 |
Hb_000540_100 |
0.103240285 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 15 |
Hb_000701_030 |
0.1032925765 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 16 |
Hb_004030_060 |
0.1033422375 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 17 |
Hb_007897_030 |
0.1040970922 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR2 [Jatropha curcas] |
| 18 |
Hb_009476_120 |
0.1055235806 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 19 |
Hb_001511_180 |
0.1061518721 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 20 |
Hb_000167_120 |
0.1063672223 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |