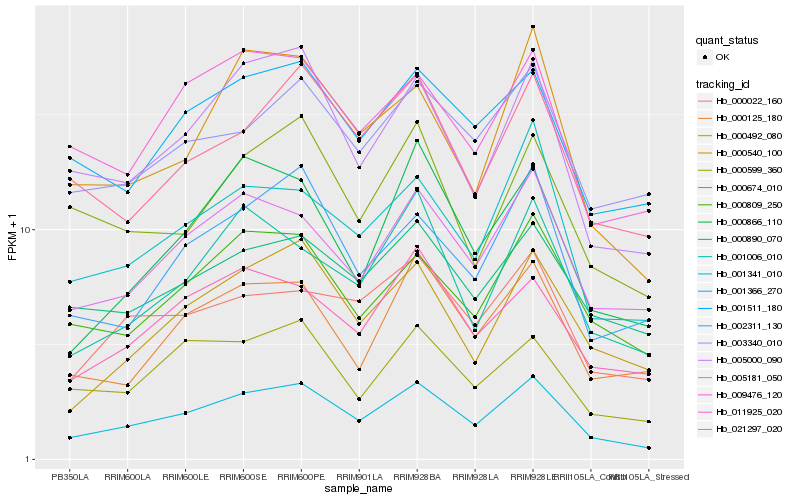
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001366_270 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 2 |
Hb_005000_090 |
0.0841854342 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 3 |
Hb_005181_050 |
0.0866134713 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 4 |
Hb_000809_250 |
0.0928638396 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 5 |
Hb_001341_010 |
0.0929898904 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001511_180 |
0.0962110775 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 7 |
Hb_000540_100 |
0.0981577121 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 8 |
Hb_011925_020 |
0.0982444387 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_000890_070 |
0.0990349705 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_009476_120 |
0.0994486345 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 11 |
Hb_002311_130 |
0.1004637481 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_021297_020 |
0.10137657 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 13 |
Hb_000492_080 |
0.1023780775 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 [Jatropha curcas] |
| 14 |
Hb_000599_360 |
0.1024591821 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000125_180 |
0.1025105816 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 16 |
Hb_000022_160 |
0.1029600115 |
- |
- |
- |
| 17 |
Hb_003340_010 |
0.1046251148 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 18 |
Hb_001006_010 |
0.1050410603 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 19 |
Hb_000674_010 |
0.107722942 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000866_110 |
0.1079428693 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |