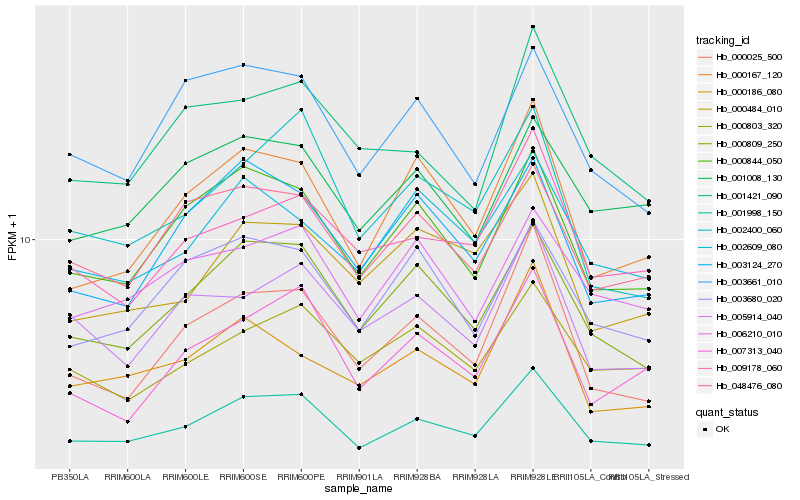
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001998_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 2 |
Hb_000809_250 |
0.0707271043 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 3 |
Hb_000844_050 |
0.0756569421 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 4 |
Hb_000167_120 |
0.0830298424 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 5 |
Hb_000025_500 |
0.086970575 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 6 |
Hb_007313_040 |
0.0927014393 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 7 |
Hb_048476_080 |
0.09298039 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 8 |
Hb_002400_060 |
0.0944570772 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 9 |
Hb_009178_060 |
0.0971204692 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_003124_270 |
0.0993542449 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 11 |
Hb_006210_010 |
0.0996202574 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 12 |
Hb_003680_020 |
0.1003342955 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_003661_010 |
0.1009075966 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001008_130 |
0.1014700848 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 15 |
Hb_000803_320 |
0.1016291246 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 16 |
Hb_000484_010 |
0.1019637217 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |
| 17 |
Hb_001421_090 |
0.1028192104 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 18 |
Hb_005914_040 |
0.1032000002 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 19 |
Hb_002609_080 |
0.1033507814 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000186_080 |
0.10509526 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |