| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
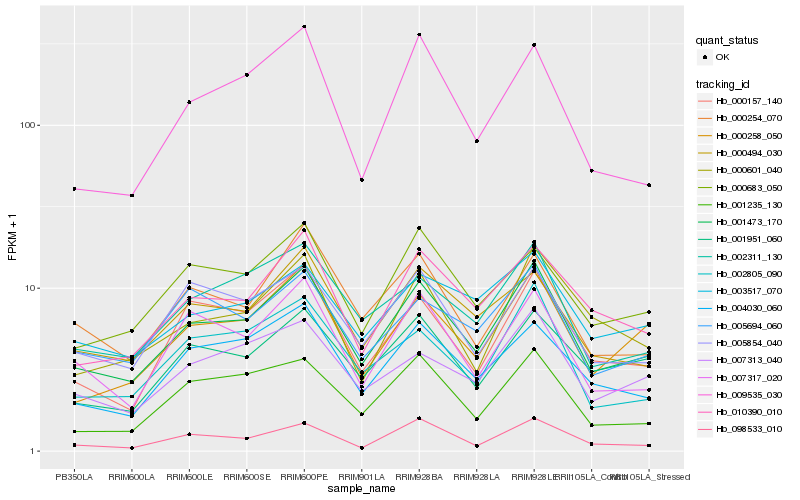
Hb_001473_170 |
0.0 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 2 |
Hb_002311_130 |
0.091400481 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_010390_010 |
0.0918553727 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 4 |
Hb_000258_050 |
0.0929433569 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000494_030 |
0.0941895276 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 6 |
Hb_004030_060 |
0.0949595337 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 7 |
Hb_007313_040 |
0.0958491865 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 8 |
Hb_002805_090 |
0.0971600554 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 9 |
Hb_000157_140 |
0.0973580635 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 10 |
Hb_001235_130 |
0.0984946965 |
- |
- |
- |
| 11 |
Hb_001951_060 |
0.0991906976 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 12 |
Hb_005854_040 |
0.1002631031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007317_020 |
0.1010826101 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000683_050 |
0.102468401 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 15 |
Hb_003517_070 |
0.1042399987 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 16 |
Hb_009535_030 |
0.1045801454 |
- |
- |
CP2 [Hevea brasiliensis] |
| 17 |
Hb_005694_060 |
0.1057261702 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 18 |
Hb_000254_070 |
0.1059388784 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 19 |
Hb_098533_010 |
0.108635362 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 20 |
Hb_000601_040 |
0.10907535 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |