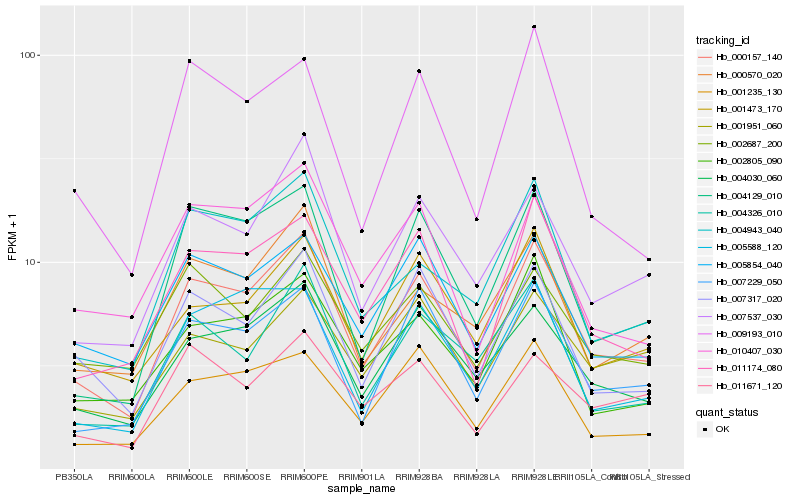
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000157_140 |
0.0 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 2 |
Hb_004030_060 |
0.0803891558 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 3 |
Hb_007537_030 |
0.0896445976 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007229_050 |
0.0903546011 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007317_020 |
0.0927827097 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011174_080 |
0.0934197885 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 7 |
Hb_000570_020 |
0.0962185625 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 8 |
Hb_004129_010 |
0.0963874933 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 9 |
Hb_005854_040 |
0.0967323534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001473_170 |
0.0973580635 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 11 |
Hb_001235_130 |
0.1005868541 |
- |
- |
- |
| 12 |
Hb_004943_040 |
0.1010637348 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004326_010 |
0.1052910128 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_002805_090 |
0.1075837076 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 15 |
Hb_001951_060 |
0.1125099766 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 16 |
Hb_005588_120 |
0.1133571695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002687_200 |
0.1133906069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010407_030 |
0.1136216125 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 19 |
Hb_009193_010 |
0.113845246 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 20 |
Hb_011671_120 |
0.1140471881 |
- |
- |
hypothetical protein JCGZ_26275 [Jatropha curcas] |