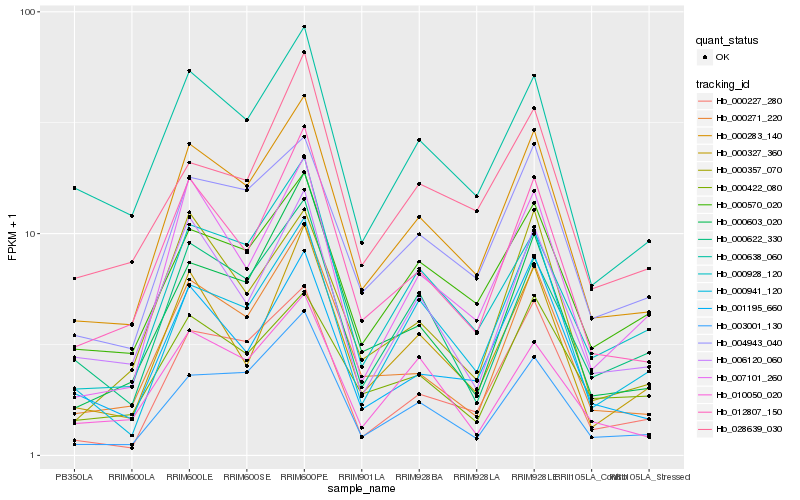
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000283_140 |
0.0 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_012807_150 |
0.0712624636 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 3 |
Hb_004943_040 |
0.0727126501 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000638_060 |
0.0885884704 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 5 |
Hb_000570_020 |
0.0895955439 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 6 |
Hb_000928_120 |
0.0908861901 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 7 |
Hb_000271_220 |
0.0941060392 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_001195_660 |
0.1018937679 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 9 |
Hb_006120_060 |
0.1019818865 |
- |
- |
O-acetyltransferase family protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_000422_080 |
0.1022432878 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 11 |
Hb_007101_260 |
0.102574527 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 12 |
Hb_003001_130 |
0.1042199084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000603_020 |
0.104443955 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000622_330 |
0.1072685934 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 15 |
Hb_000327_360 |
0.1084848126 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_000941_120 |
0.1086223735 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 17 |
Hb_000357_070 |
0.1091193663 |
- |
- |
- |
| 18 |
Hb_010050_020 |
0.1102076363 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 19 |
Hb_028639_030 |
0.1106836124 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 20 |
Hb_000227_280 |
0.1134699808 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |