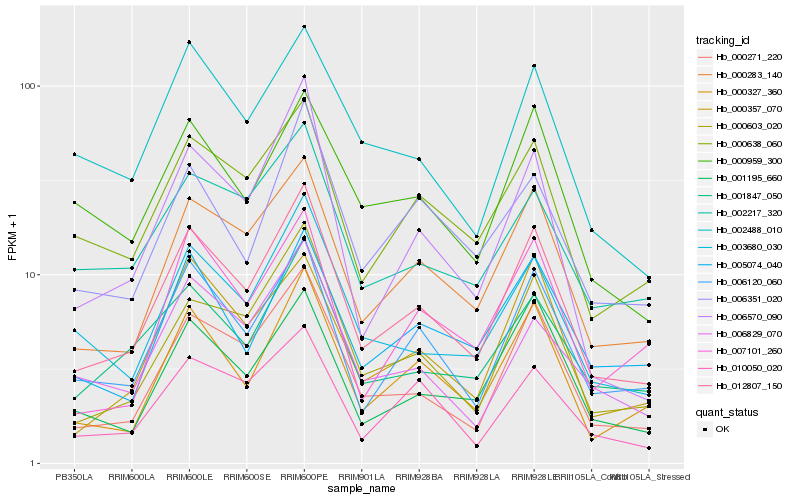
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012807_150 |
0.0 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 2 |
Hb_000283_140 |
0.0712624636 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000271_220 |
0.0750499481 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 4 |
Hb_006120_060 |
0.0851837312 |
- |
- |
O-acetyltransferase family protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_000603_020 |
0.0904816424 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000327_360 |
0.0930095235 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000638_060 |
0.1007394068 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 8 |
Hb_003680_030 |
0.102774228 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 9 |
Hb_001847_050 |
0.1052579832 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 10 |
Hb_001195_660 |
0.1066982009 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 11 |
Hb_000357_070 |
0.1104525081 |
- |
- |
- |
| 12 |
Hb_010050_020 |
0.1140134195 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 13 |
Hb_002217_320 |
0.1169895982 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006570_090 |
0.1174940599 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 15 |
Hb_005074_040 |
0.1177776973 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_002488_010 |
0.1184136266 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 17 |
Hb_006351_020 |
0.1186473333 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 18 |
Hb_006829_070 |
0.1200068025 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 19 |
Hb_000959_300 |
0.1205376648 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 20 |
Hb_007101_260 |
0.1205449304 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |