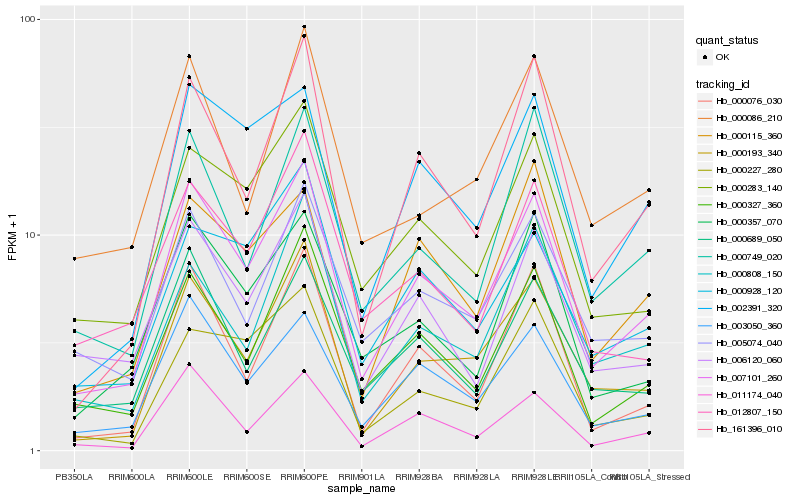
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007101_260 |
0.0 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 2 |
Hb_003050_360 |
0.0890158258 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000327_360 |
0.0974630584 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_161396_010 |
0.0994640886 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |
| 5 |
Hb_000193_340 |
0.1000573413 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000283_140 |
0.102574527 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_006120_060 |
0.1079056588 |
- |
- |
O-acetyltransferase family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_002391_320 |
0.1103233187 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 9 |
Hb_000689_050 |
0.1111726627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011174_040 |
0.1119060257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000076_030 |
0.1121607603 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000357_070 |
0.1131589118 |
- |
- |
- |
| 13 |
Hb_000749_020 |
0.1150510745 |
- |
- |
Nucleotide-diphospho-sugar transferases superfamily protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_005074_040 |
0.115304242 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000928_120 |
0.1195215544 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 16 |
Hb_012807_150 |
0.1205449304 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 17 |
Hb_000808_150 |
0.1247304516 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 18 |
Hb_000227_280 |
0.126040909 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 19 |
Hb_000115_360 |
0.1266656837 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000086_210 |
0.1282697706 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |