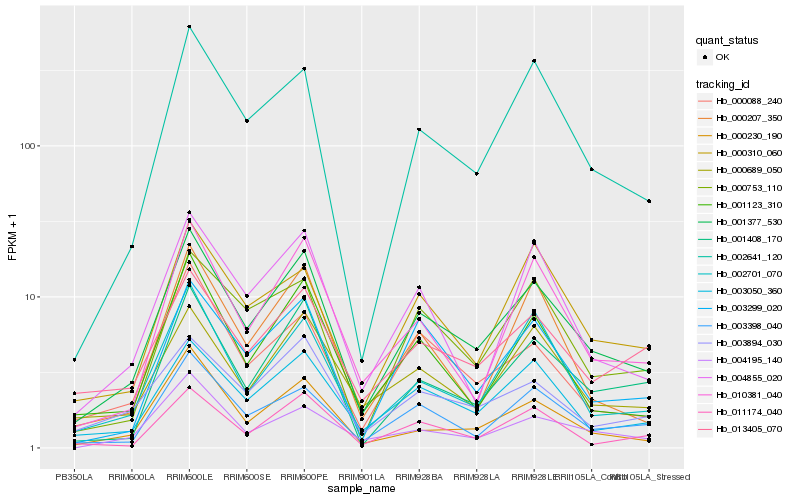
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_530 |
0.0 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 2 |
Hb_003050_360 |
0.1036154082 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002641_120 |
0.1090027 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 4 |
Hb_003398_040 |
0.1143574651 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001123_310 |
0.1203565518 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 6 |
Hb_003299_020 |
0.1213230321 |
- |
- |
PREDICTED: uncharacterized protein LOC105649783 [Jatropha curcas] |
| 7 |
Hb_001408_170 |
0.1216489562 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 2 [Jatropha curcas] |
| 8 |
Hb_010381_040 |
0.1217410113 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 9 |
Hb_003894_030 |
0.1225251853 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 10 |
Hb_000689_050 |
0.1244652429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000230_190 |
0.1249214687 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004855_020 |
0.1275081181 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 13 |
Hb_000753_110 |
0.1275698709 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X6 [Jatropha curcas] |
| 14 |
Hb_000088_240 |
0.1276185246 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 15 |
Hb_002701_070 |
0.1286782933 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 16 |
Hb_011174_040 |
0.130752958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000310_060 |
0.1321200345 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 18 |
Hb_013405_070 |
0.1354750054 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 19 |
Hb_004195_140 |
0.135843202 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 20 |
Hb_000207_350 |
0.1364319038 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |