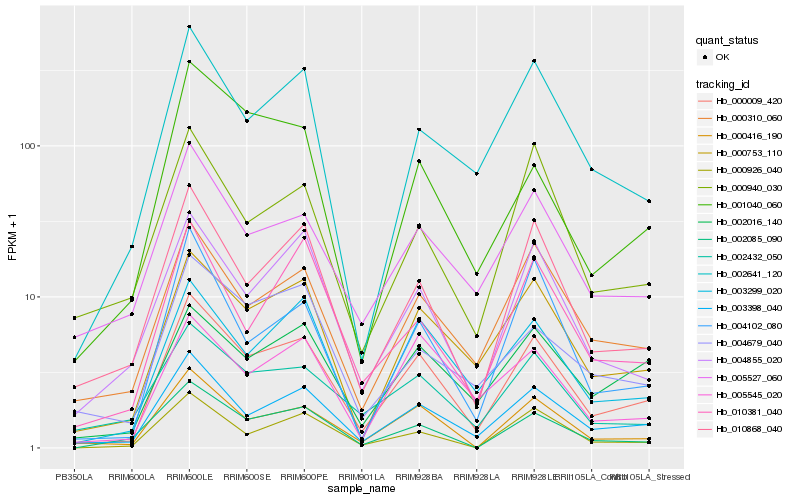
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_420 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639614 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003398_040 |
0.0926236784 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000753_110 |
0.1088366585 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X6 [Jatropha curcas] |
| 4 |
Hb_000416_190 |
0.1169698844 |
- |
- |
- |
| 5 |
Hb_000310_060 |
0.1244472657 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 6 |
Hb_010868_040 |
0.124649945 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 7 |
Hb_003299_020 |
0.1266945021 |
- |
- |
PREDICTED: uncharacterized protein LOC105649783 [Jatropha curcas] |
| 8 |
Hb_004855_020 |
0.1375074794 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 9 |
Hb_005527_060 |
0.1398982611 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 10 |
Hb_002641_120 |
0.1411131751 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 11 |
Hb_002016_140 |
0.1436307097 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 12 |
Hb_002085_090 |
0.1437090342 |
- |
- |
- |
| 13 |
Hb_004102_080 |
0.1475654947 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 14 |
Hb_002432_050 |
0.1477819844 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 15 |
Hb_000926_040 |
0.1478368505 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH5 [Jatropha curcas] |
| 16 |
Hb_005545_020 |
0.1499526515 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |
| 17 |
Hb_010381_040 |
0.1551643793 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 18 |
Hb_004679_040 |
0.1552693026 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 19 |
Hb_001040_060 |
0.155573283 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 20 |
Hb_000940_030 |
0.1569487393 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |