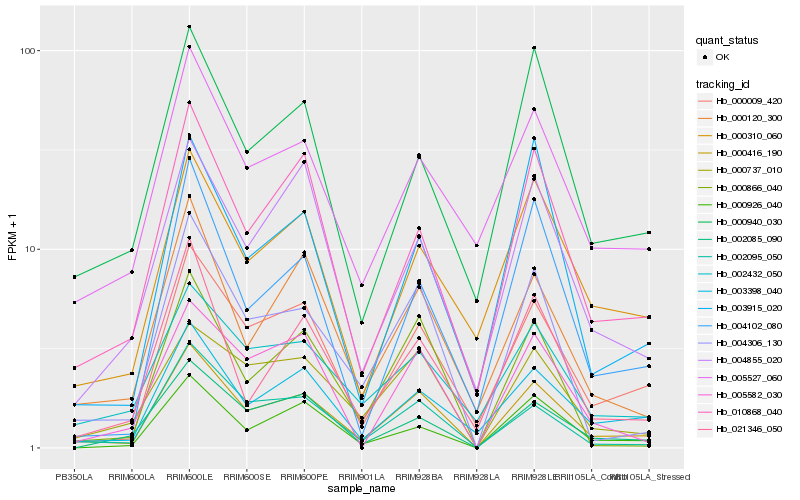
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000416_190 |
0.0 |
- |
- |
- |
| 2 |
Hb_000009_420 |
0.1169698844 |
- |
- |
PREDICTED: uncharacterized protein LOC105639614 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004306_130 |
0.1214015431 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010868_040 |
0.1277430893 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 5 |
Hb_002095_050 |
0.130382008 |
- |
- |
PREDICTED: uncharacterized protein LOC104447479 [Eucalyptus grandis] |
| 6 |
Hb_003398_040 |
0.136830921 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000120_300 |
0.137518625 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X1 [Eucalyptus grandis] |
| 8 |
Hb_000737_010 |
0.138957719 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 9 |
Hb_000926_040 |
0.1444449533 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH5 [Jatropha curcas] |
| 10 |
Hb_005582_030 |
0.1485255632 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A-like [Jatropha curcas] |
| 11 |
Hb_002085_090 |
0.1488920245 |
- |
- |
- |
| 12 |
Hb_000866_040 |
0.1520864221 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 13 |
Hb_021346_050 |
0.1522250634 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 14 |
Hb_002432_050 |
0.1522596085 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 15 |
Hb_004855_020 |
0.1529658281 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 16 |
Hb_004102_080 |
0.1531620718 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 17 |
Hb_000940_030 |
0.154323429 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000310_060 |
0.1558789286 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 19 |
Hb_005527_060 |
0.1564970795 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 20 |
Hb_003915_020 |
0.1611339551 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |