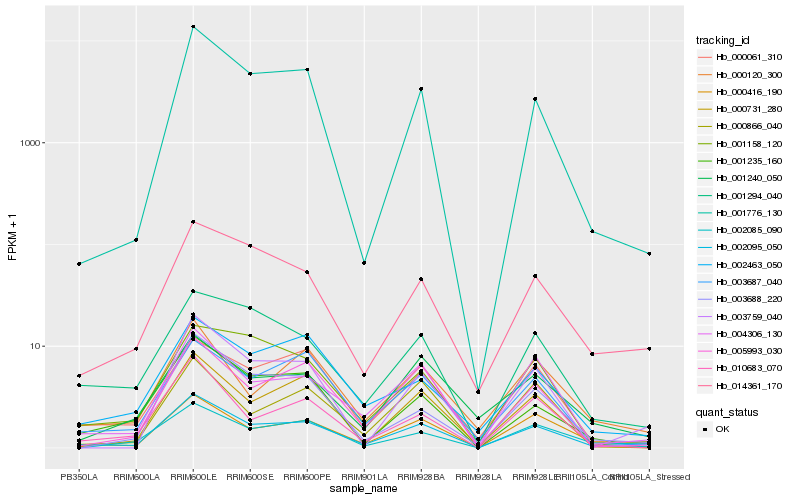
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002095_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104447479 [Eucalyptus grandis] |
| 2 |
Hb_001776_130 |
0.1182330209 |
- |
- |
metallothionein [Hevea brasiliensis] |
| 3 |
Hb_004306_130 |
0.1232010141 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005993_030 |
0.1293115422 |
- |
- |
putative senescence-associated protein SAG102 [Populus trichocarpa] |
| 5 |
Hb_000416_190 |
0.130382008 |
- |
- |
- |
| 6 |
Hb_003687_040 |
0.1362953074 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 7 |
Hb_014361_170 |
0.1373323263 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 37 [Jatropha curcas] |
| 8 |
Hb_000731_280 |
0.1406154719 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000061_310 |
0.1474339374 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 10 |
Hb_010683_070 |
0.1488791189 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001235_160 |
0.1489522383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001294_040 |
0.1506478269 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 13 |
Hb_003688_220 |
0.1522135775 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_000120_300 |
0.1531993481 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X1 [Eucalyptus grandis] |
| 15 |
Hb_002463_050 |
0.1534491747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003759_040 |
0.1568250744 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 17 |
Hb_002085_090 |
0.1586369085 |
- |
- |
- |
| 18 |
Hb_001158_120 |
0.1624164888 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000866_040 |
0.1630686051 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 20 |
Hb_001240_050 |
0.1631575178 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Jatropha curcas] |