| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
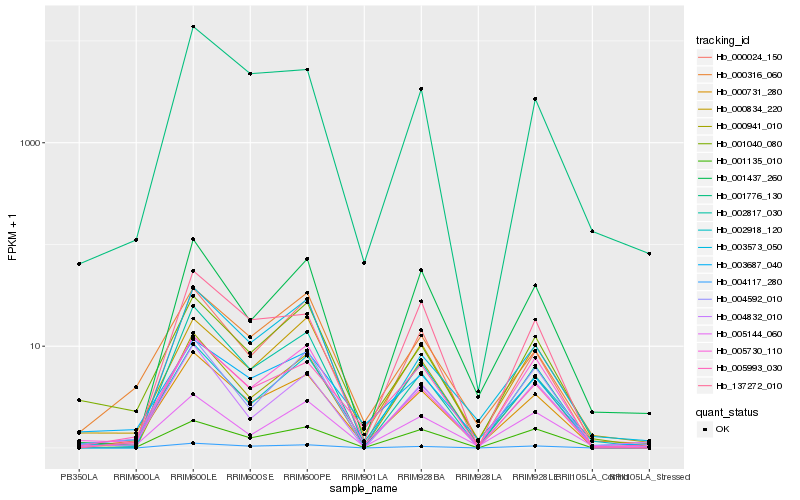
Hb_000731_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005993_030 |
0.0917125486 |
- |
- |
putative senescence-associated protein SAG102 [Populus trichocarpa] |
| 3 |
Hb_000941_010 |
0.108077797 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 4 |
Hb_002918_120 |
0.1109051769 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_004117_280 |
0.1196681455 |
- |
- |
hypothetical protein JCGZ_00361 [Jatropha curcas] |
| 6 |
Hb_000024_150 |
0.1200042543 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 7 |
Hb_001437_260 |
0.122726552 |
- |
- |
PREDICTED: protein FAM98B-like [Jatropha curcas] |
| 8 |
Hb_003687_040 |
0.1232983734 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 9 |
Hb_001135_010 |
0.1238112 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 10 |
Hb_001040_080 |
0.1264116098 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 11 |
Hb_000834_220 |
0.1264245696 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 12 |
Hb_002817_030 |
0.1280295002 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 13 |
Hb_001776_130 |
0.1280920956 |
- |
- |
metallothionein [Hevea brasiliensis] |
| 14 |
Hb_004832_010 |
0.1281545482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004592_010 |
0.1285657247 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b-like [Jatropha curcas] |
| 16 |
Hb_137272_010 |
0.1313241204 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 17 |
Hb_005730_110 |
0.1316760842 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 18 |
Hb_000316_060 |
0.1320414262 |
- |
- |
hypothetical protein CISIN_1g017081mg [Citrus sinensis] |
| 19 |
Hb_005144_060 |
0.1330818819 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 20 |
Hb_003573_050 |
0.1392376971 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |