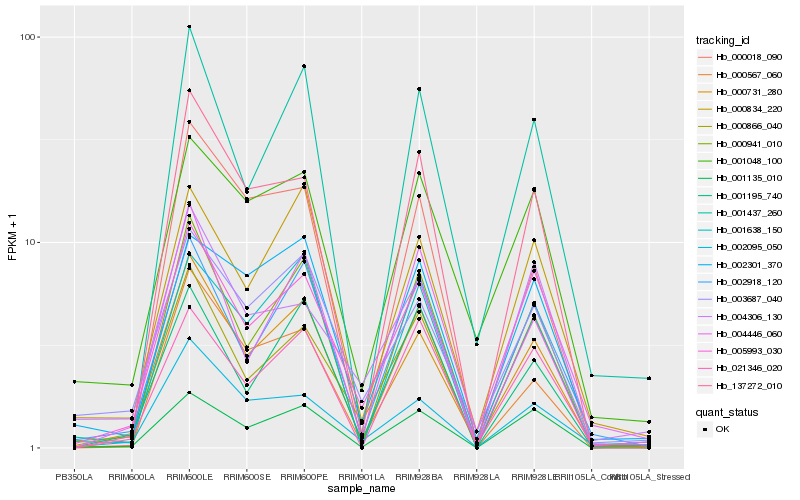
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005993_030 |
0.0 |
- |
- |
putative senescence-associated protein SAG102 [Populus trichocarpa] |
| 2 |
Hb_000731_280 |
0.0917125486 |
- |
- |
PREDICTED: uncharacterized protein LOC105649351 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000866_040 |
0.1149469182 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 4 |
Hb_002918_120 |
0.1150909788 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_003687_040 |
0.1154939271 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 6 |
Hb_000941_010 |
0.1262066674 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 7 |
Hb_001195_740 |
0.1292078216 |
transcription factor |
TF Family: LIM |
PREDICTED: protein DA1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002095_050 |
0.1293115422 |
- |
- |
PREDICTED: uncharacterized protein LOC104447479 [Eucalyptus grandis] |
| 9 |
Hb_000567_060 |
0.1315399324 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |
| 10 |
Hb_137272_010 |
0.1345755819 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 11 |
Hb_001135_010 |
0.1395844877 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 12 |
Hb_004446_060 |
0.1437631917 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 13 |
Hb_001437_260 |
0.143990994 |
- |
- |
PREDICTED: protein FAM98B-like [Jatropha curcas] |
| 14 |
Hb_000834_220 |
0.1455018337 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 15 |
Hb_002301_370 |
0.1501340877 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 16 |
Hb_021346_020 |
0.1502008238 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 17 |
Hb_001638_150 |
0.1507412925 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 18 |
Hb_004306_130 |
0.1523558456 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001048_100 |
0.1524011216 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 20 |
Hb_000018_090 |
0.1531044294 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |