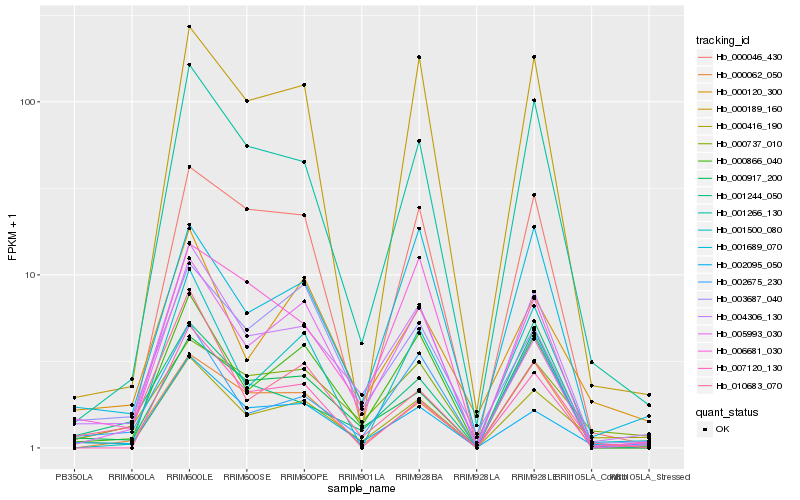
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004306_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000866_040 |
0.1011847035 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 3 |
Hb_000416_190 |
0.1214015431 |
- |
- |
- |
| 4 |
Hb_001266_130 |
0.1226965714 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 5 |
Hb_002095_050 |
0.1232010141 |
- |
- |
PREDICTED: uncharacterized protein LOC104447479 [Eucalyptus grandis] |
| 6 |
Hb_002675_230 |
0.1496405206 |
- |
- |
PREDICTED: glutathione S-transferase L3-like [Populus euphratica] |
| 7 |
Hb_005993_030 |
0.1523558456 |
- |
- |
putative senescence-associated protein SAG102 [Populus trichocarpa] |
| 8 |
Hb_000737_010 |
0.1555601986 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 9 |
Hb_001689_070 |
0.1576628664 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_010683_070 |
0.1655942213 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000189_160 |
0.1689348021 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 12 |
Hb_006681_030 |
0.1707030418 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 13 |
Hb_000917_200 |
0.1712627562 |
- |
- |
hypothetical protein PRUPE_ppa010555mg [Prunus persica] |
| 14 |
Hb_000120_300 |
0.1743308434 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X1 [Eucalyptus grandis] |
| 15 |
Hb_001500_080 |
0.1762435139 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001244_050 |
0.1765274326 |
- |
- |
PREDICTED: uncharacterized protein LOC105643334 [Jatropha curcas] |
| 17 |
Hb_000046_430 |
0.1770690622 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 18 |
Hb_000062_050 |
0.1773166894 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_007120_130 |
0.1779183698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003687_040 |
0.1779784968 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |