| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
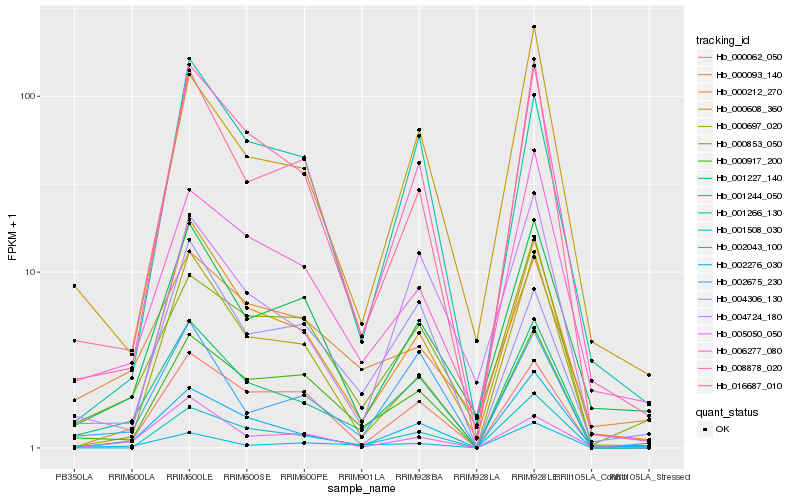
Hb_001244_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643334 [Jatropha curcas] |
| 2 |
Hb_002276_030 |
0.1208185579 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 3 |
Hb_008878_020 |
0.1331051426 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 4 |
Hb_000917_200 |
0.1375241845 |
- |
- |
hypothetical protein PRUPE_ppa010555mg [Prunus persica] |
| 5 |
Hb_016687_010 |
0.1466833164 |
- |
- |
PREDICTED: uncharacterized protein LOC105637401 [Jatropha curcas] |
| 6 |
Hb_002675_230 |
0.1550729503 |
- |
- |
PREDICTED: glutathione S-transferase L3-like [Populus euphratica] |
| 7 |
Hb_000212_270 |
0.1572811652 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 8 |
Hb_006277_080 |
0.158182208 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 9 |
Hb_000853_050 |
0.1601202582 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_001227_140 |
0.1650780858 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 11 |
Hb_000608_360 |
0.1662948692 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 12 |
Hb_004724_180 |
0.1713992308 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 13 |
Hb_000062_050 |
0.1722906432 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_001508_030 |
0.1725629787 |
- |
- |
hypothetical protein VITISV_029808 [Vitis vinifera] |
| 15 |
Hb_002043_100 |
0.1750684861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000093_140 |
0.1751623982 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001266_130 |
0.1760576071 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 18 |
Hb_004306_130 |
0.1765274326 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000697_020 |
0.1768926171 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_005050_050 |
0.1771059444 |
- |
- |
PREDICTED: wall-associated receptor kinase 5-like [Glycine max] |