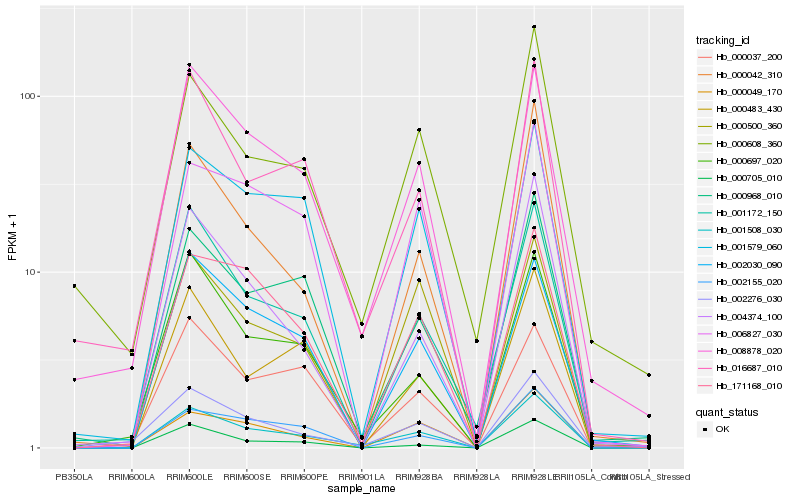
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001508_030 |
0.0 |
- |
- |
hypothetical protein VITISV_029808 [Vitis vinifera] |
| 2 |
Hb_008878_020 |
0.1137225509 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 3 |
Hb_000049_170 |
0.1259940495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_171168_010 |
0.129453879 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_016687_010 |
0.1302156484 |
- |
- |
PREDICTED: uncharacterized protein LOC105637401 [Jatropha curcas] |
| 6 |
Hb_000968_010 |
0.1309390227 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000042_310 |
0.1322089959 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 8 |
Hb_001172_150 |
0.132360805 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 9 |
Hb_000697_020 |
0.1323916315 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_001579_060 |
0.1334974222 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 11 |
Hb_002155_020 |
0.1335920744 |
- |
- |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 12 |
Hb_006827_030 |
0.1367809977 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 13 |
Hb_004374_100 |
0.1384641931 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |
| 14 |
Hb_002030_090 |
0.1393528352 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 15 |
Hb_002276_030 |
0.1398638382 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 16 |
Hb_000705_010 |
0.1443497932 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 17 |
Hb_000483_430 |
0.1477996897 |
- |
- |
- |
| 18 |
Hb_000608_360 |
0.1478673046 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 19 |
Hb_000500_360 |
0.1497395181 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000037_200 |
0.1533627588 |
- |
- |
RNA binding protein, putative [Ricinus communis] |