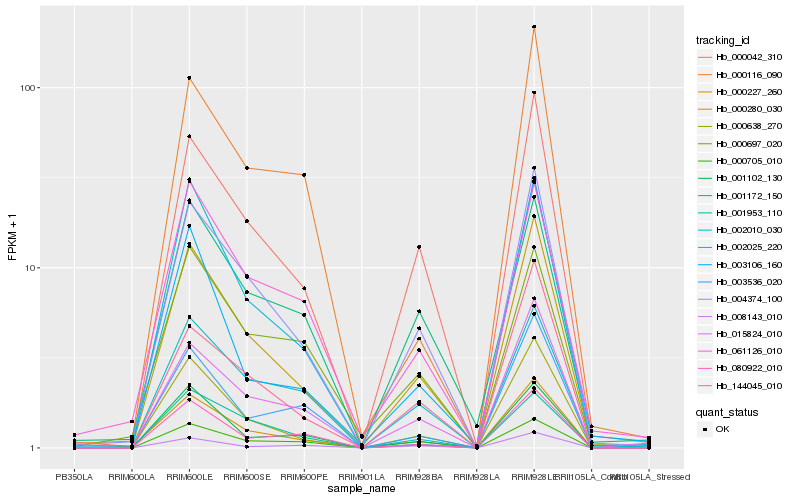
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000280_030 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 2 |
Hb_004374_100 |
0.0557184439 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |
| 3 |
Hb_000638_270 |
0.0625903007 |
- |
- |
- |
| 4 |
Hb_000042_310 |
0.0683493052 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 5 |
Hb_015824_010 |
0.1004331939 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 6 |
Hb_000705_010 |
0.1051855533 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 7 |
Hb_002025_220 |
0.1115088298 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000227_260 |
0.1152506222 |
- |
- |
PREDICTED: RAN GTPase-activating protein 2 [Jatropha curcas] |
| 9 |
Hb_144045_010 |
0.1166861471 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 10 |
Hb_080922_010 |
0.127642658 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_003536_020 |
0.1360710243 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 12 |
Hb_003106_160 |
0.1361384921 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 13 |
Hb_001102_130 |
0.1370855713 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 14 |
Hb_061126_010 |
0.1376039884 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 15 |
Hb_008143_010 |
0.1392572554 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 16 |
Hb_000116_090 |
0.1433627758 |
- |
- |
catalase [Hevea brasiliensis] |
| 17 |
Hb_001172_150 |
0.1440583624 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 18 |
Hb_002010_030 |
0.1492299854 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 19 |
Hb_001953_110 |
0.1510907652 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 20 |
Hb_000697_020 |
0.1517499259 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |