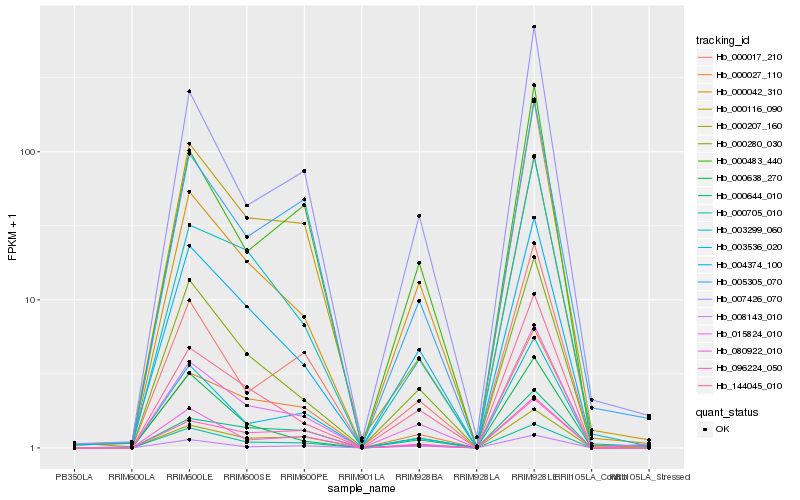
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015824_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 2 |
Hb_000042_310 |
0.0875054581 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 3 |
Hb_004374_100 |
0.0954594972 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |
| 4 |
Hb_144045_010 |
0.0985177125 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 5 |
Hb_000280_030 |
0.1004331939 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 6 |
Hb_000027_110 |
0.1041733098 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 7 |
Hb_000207_160 |
0.1099751293 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Populus euphratica] |
| 8 |
Hb_003536_020 |
0.1105339457 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 9 |
Hb_000705_010 |
0.1136343706 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 10 |
Hb_005305_070 |
0.1154973512 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 11 |
Hb_000116_090 |
0.1174610102 |
- |
- |
catalase [Hevea brasiliensis] |
| 12 |
Hb_007426_070 |
0.1176516606 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 13 |
Hb_000483_440 |
0.1203395821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000644_010 |
0.1215733299 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_008143_010 |
0.1220830535 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 16 |
Hb_003299_060 |
0.1266619768 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 17 |
Hb_080922_010 |
0.1282821877 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 18 |
Hb_000017_210 |
0.1320992478 |
- |
- |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 19 |
Hb_096224_050 |
0.1330156532 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 20 |
Hb_000638_270 |
0.1362030874 |
- |
- |
- |