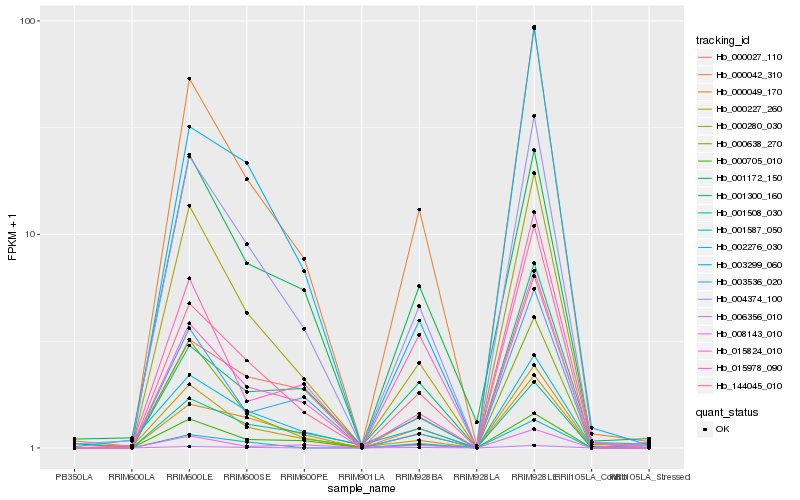
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_310 |
0.0 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 2 |
Hb_004374_100 |
0.0473118225 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |
| 3 |
Hb_000280_030 |
0.0683493052 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 4 |
Hb_015824_010 |
0.0875054581 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 5 |
Hb_144045_010 |
0.0889162816 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 6 |
Hb_000638_270 |
0.1152985898 |
- |
- |
- |
| 7 |
Hb_000705_010 |
0.1199699233 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 8 |
Hb_008143_010 |
0.12509734 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 9 |
Hb_001508_030 |
0.1322089959 |
- |
- |
hypothetical protein VITISV_029808 [Vitis vinifera] |
| 10 |
Hb_001172_150 |
0.13316343 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 11 |
Hb_000227_260 |
0.1346711517 |
- |
- |
PREDICTED: RAN GTPase-activating protein 2 [Jatropha curcas] |
| 12 |
Hb_015978_090 |
0.142111853 |
- |
- |
hypothetical protein POPTR_0014s11700g [Populus trichocarpa] |
| 13 |
Hb_000027_110 |
0.1435661639 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 14 |
Hb_003299_060 |
0.1447198804 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 15 |
Hb_006356_010 |
0.1467264945 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 16 |
Hb_001587_050 |
0.1491980718 |
- |
- |
- |
| 17 |
Hb_000049_170 |
0.1493031869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003536_020 |
0.1510797513 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 19 |
Hb_002276_030 |
0.1512254992 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 20 |
Hb_001300_160 |
0.1522953074 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |