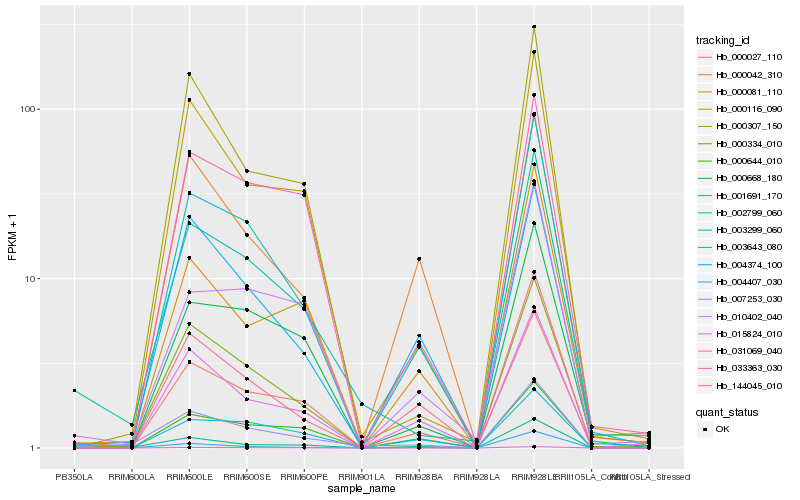
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003299_060 |
0.0 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 2 |
Hb_144045_010 |
0.0962734735 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 3 |
Hb_000027_110 |
0.0993448212 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 4 |
Hb_000668_180 |
0.1126406892 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 5 |
Hb_000334_010 |
0.1195884189 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 6 |
Hb_015824_010 |
0.1266619768 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 7 |
Hb_000116_090 |
0.1309082779 |
- |
- |
catalase [Hevea brasiliensis] |
| 8 |
Hb_004374_100 |
0.1379874807 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |
| 9 |
Hb_003643_080 |
0.1383732012 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 10 |
Hb_001691_170 |
0.13876202 |
- |
- |
PREDICTED: callose synthase 12-like [Glycine max] |
| 11 |
Hb_002799_060 |
0.14391193 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_007253_030 |
0.1444596929 |
- |
- |
PREDICTED: 33 kDa ribonucleoprotein, chloroplastic-like [Populus euphratica] |
| 13 |
Hb_000042_310 |
0.1447198804 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 14 |
Hb_000644_010 |
0.144886095 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000081_110 |
0.1454253499 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 16 |
Hb_010402_040 |
0.1473623461 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 17 |
Hb_033363_030 |
0.1497320609 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 18 |
Hb_031069_040 |
0.1499829503 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_000307_150 |
0.15086094 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004407_030 |
0.151685281 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |