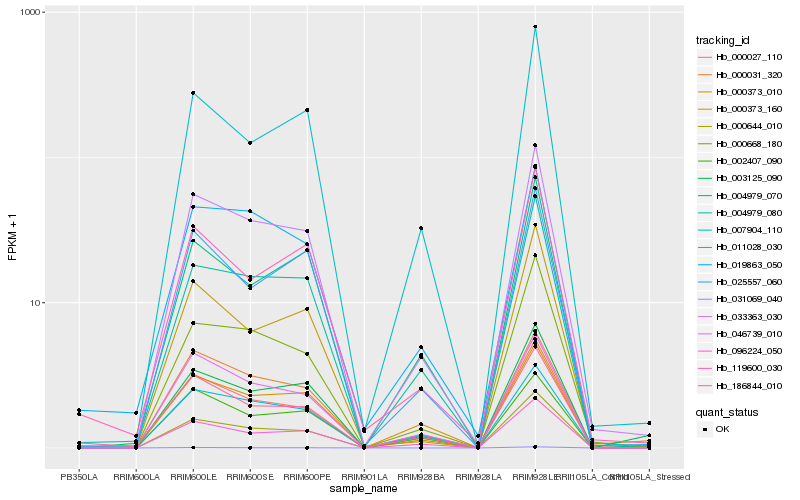
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033363_030 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 2 |
Hb_096224_050 |
0.0687699783 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 3 |
Hb_004979_080 |
0.075932204 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_000027_110 |
0.0775308851 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 5 |
Hb_000668_180 |
0.0793927191 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 6 |
Hb_046739_010 |
0.0865589571 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 7 |
Hb_011028_030 |
0.0872363915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_025557_060 |
0.0922395614 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_007904_110 |
0.0983144469 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_000644_010 |
0.0989636947 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003125_090 |
0.1011740851 |
- |
- |
- |
| 12 |
Hb_186844_010 |
0.101515922 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_019863_050 |
0.1015930727 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 14 |
Hb_002407_090 |
0.102579025 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 15 |
Hb_000373_010 |
0.1027907215 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000373_160 |
0.1038402266 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 17 |
Hb_004979_070 |
0.1042627994 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |
| 18 |
Hb_031069_040 |
0.1059424371 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_119600_030 |
0.1070840617 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 20 |
Hb_000031_320 |
0.1077518348 |
- |
- |
unnamed protein product [Vitis vinifera] |