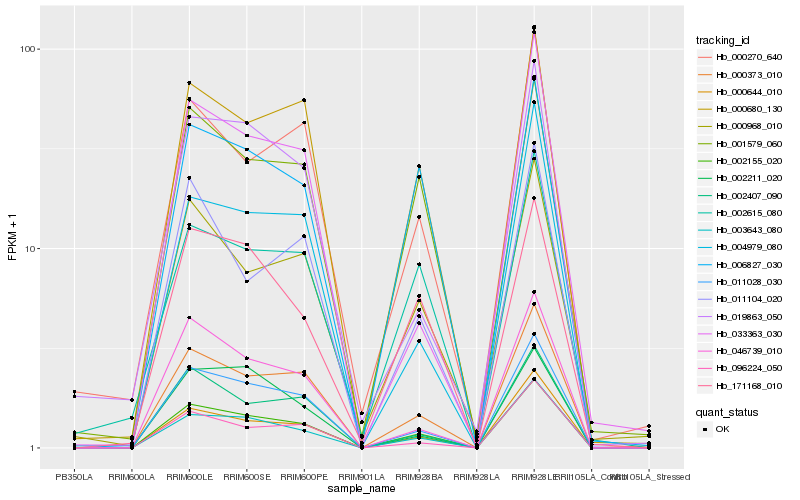
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002155_020 |
0.0 |
- |
- |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 2 |
Hb_011028_030 |
0.0603889814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000373_010 |
0.0679544351 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_003643_080 |
0.071956807 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 5 |
Hb_000644_010 |
0.075337025 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000680_130 |
0.0861728261 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 7 |
Hb_002407_090 |
0.0976546756 |
- |
- |
hypothetical protein JCGZ_00747 [Jatropha curcas] |
| 8 |
Hb_006827_030 |
0.1036369977 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 9 |
Hb_046739_010 |
0.1047166677 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001579_060 |
0.1051772074 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 11 |
Hb_000968_010 |
0.1078752145 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_171168_010 |
0.1089752589 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_096224_050 |
0.110899415 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 14 |
Hb_002615_080 |
0.120892602 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 15 |
Hb_011104_020 |
0.1228478163 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 16 |
Hb_000270_640 |
0.1237796589 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 17 |
Hb_004979_080 |
0.1240411154 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_019863_050 |
0.1271910845 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 19 |
Hb_033363_030 |
0.1276849873 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002211_020 |
0.1285283747 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |