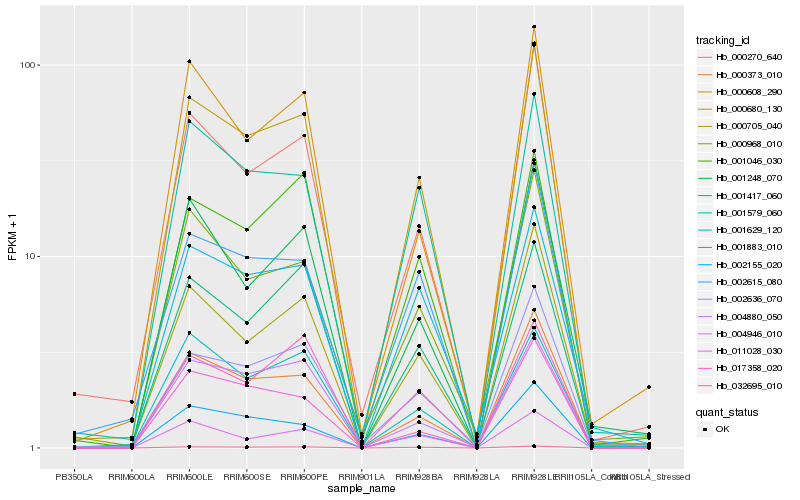
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000680_130 |
0.0 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 2 |
Hb_000373_010 |
0.0767705219 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000705_040 |
0.0792321414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002155_020 |
0.0861728261 |
- |
- |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 5 |
Hb_002636_070 |
0.0874007327 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 6 |
Hb_032695_010 |
0.087513626 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 7 |
Hb_001579_060 |
0.0878519359 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 8 |
Hb_000270_640 |
0.0943889889 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 9 |
Hb_000968_010 |
0.0944873383 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001883_010 |
0.0966056532 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 11 |
Hb_001417_060 |
0.0977786523 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 12 |
Hb_002615_080 |
0.0984159227 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 13 |
Hb_001046_030 |
0.100028469 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_004946_010 |
0.1025794374 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 15 |
Hb_004880_050 |
0.1026188897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001248_070 |
0.1089184974 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |
| 17 |
Hb_017358_020 |
0.1093086024 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |
| 18 |
Hb_011028_030 |
0.111199618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000608_290 |
0.1114063168 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 20 |
Hb_001629_120 |
0.1120377008 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |