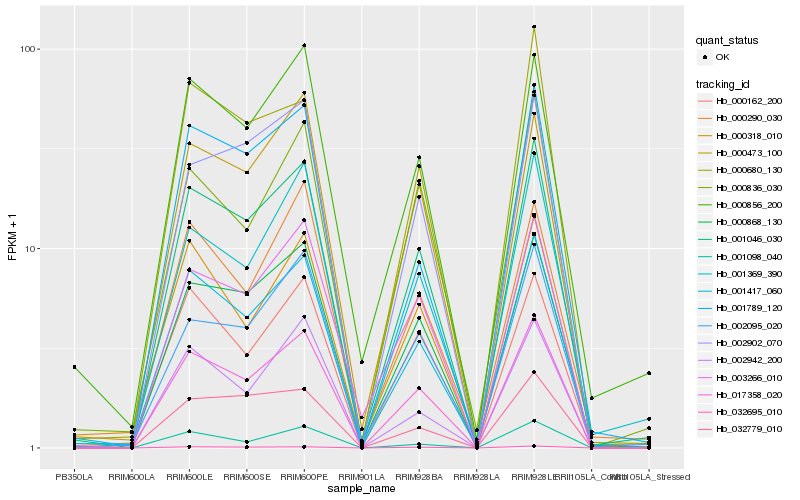
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017358_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |
| 2 |
Hb_001046_030 |
0.0460833568 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_001417_060 |
0.0556887776 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 4 |
Hb_000868_130 |
0.063756205 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 5 |
Hb_003266_010 |
0.0852221063 |
- |
- |
hypothetical protein JCGZ_21945 [Jatropha curcas] |
| 6 |
Hb_032695_010 |
0.0899295242 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 7 |
Hb_000162_200 |
0.0935136165 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000473_100 |
0.0939523389 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 9 |
Hb_002942_200 |
0.098542207 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH137 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002095_020 |
0.0990130387 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002902_070 |
0.0991481503 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 12 |
Hb_001369_390 |
0.101208583 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 13 |
Hb_001098_040 |
0.101874683 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 14 |
Hb_000318_010 |
0.1022517123 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 15 |
Hb_000836_030 |
0.1046557792 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 16 |
Hb_001789_120 |
0.1075551993 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000680_130 |
0.1093086024 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 18 |
Hb_000290_030 |
0.1093550151 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 19 |
Hb_032779_010 |
0.1102987815 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_000856_200 |
0.1113276498 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |