| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
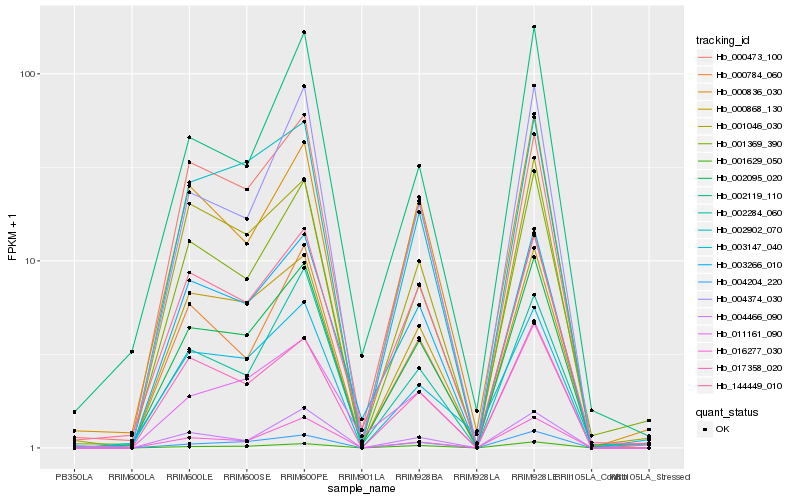
Hb_002095_020 |
0.0 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001369_390 |
0.0658374548 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 3 |
Hb_003266_010 |
0.0823579343 |
- |
- |
hypothetical protein JCGZ_21945 [Jatropha curcas] |
| 4 |
Hb_000868_130 |
0.0873420098 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 5 |
Hb_004374_030 |
0.0923506307 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 6 |
Hb_004204_220 |
0.0947302451 |
- |
- |
- |
| 7 |
Hb_000784_060 |
0.0947660758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000836_030 |
0.0956619556 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 9 |
Hb_001046_030 |
0.0974312115 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_017358_020 |
0.0990130387 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |
| 11 |
Hb_002902_070 |
0.0991211298 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 12 |
Hb_144449_010 |
0.1001106821 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 13 |
Hb_000473_100 |
0.1022509004 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 14 |
Hb_016277_030 |
0.1037397579 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 15 |
Hb_003147_040 |
0.1038524844 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 16 |
Hb_011161_090 |
0.1046208645 |
- |
- |
- |
| 17 |
Hb_002284_060 |
0.1048876442 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 18 |
Hb_002119_110 |
0.1058093475 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 19 |
Hb_001629_050 |
0.1098137854 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 20 |
Hb_004466_090 |
0.110038493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |