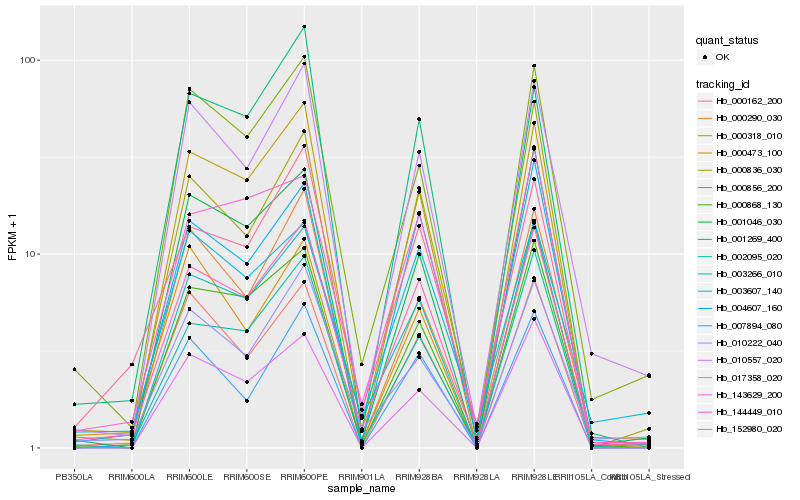
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_144449_010 |
0.0 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 2 |
Hb_000473_100 |
0.0770731186 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 3 |
Hb_003266_010 |
0.0868989275 |
- |
- |
hypothetical protein JCGZ_21945 [Jatropha curcas] |
| 4 |
Hb_010557_020 |
0.0870165919 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 5 |
Hb_004607_160 |
0.0882601111 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 6 |
Hb_152980_020 |
0.0932251746 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 7 |
Hb_000856_200 |
0.0946140885 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 8 |
Hb_010222_040 |
0.0975181803 |
- |
- |
PREDICTED: kinesin-2 [Jatropha curcas] |
| 9 |
Hb_000868_130 |
0.0991983497 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 10 |
Hb_002095_020 |
0.1001106821 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000162_200 |
0.1009336109 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 12 |
Hb_143629_200 |
0.1055958161 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 13 |
Hb_003607_140 |
0.1099868474 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 14 |
Hb_000290_030 |
0.1116204796 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 15 |
Hb_000836_030 |
0.1125479492 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 16 |
Hb_001046_030 |
0.113458371 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001269_400 |
0.1178509498 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 18 |
Hb_000318_010 |
0.1186278877 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 19 |
Hb_007894_080 |
0.119132757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_017358_020 |
0.1204232464 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |