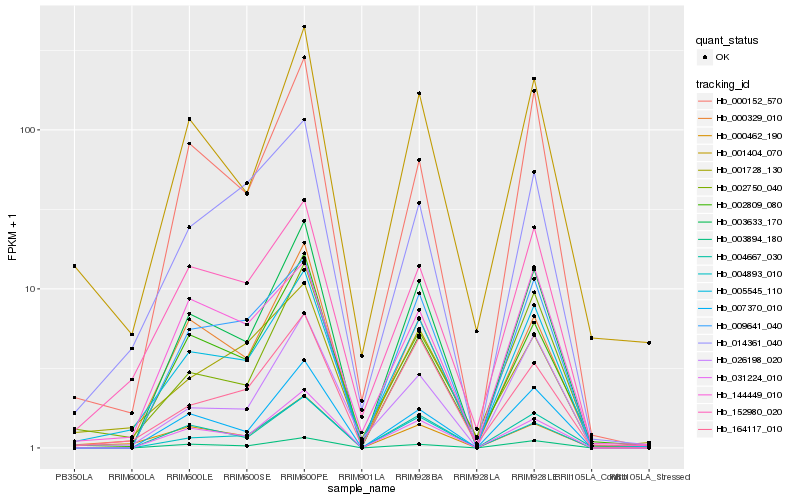
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005545_110 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 2 |
Hb_152980_020 |
0.0844307111 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 3 |
Hb_003633_170 |
0.1073283393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002750_040 |
0.1176111643 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 5 |
Hb_031224_010 |
0.1178077082 |
- |
- |
- |
| 6 |
Hb_001404_070 |
0.1185480796 |
- |
- |
sucrose synthase 4 [Hevea brasiliensis] |
| 7 |
Hb_001728_130 |
0.120075154 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_009641_040 |
0.1203843731 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 9 |
Hb_164117_010 |
0.1227069533 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 10 |
Hb_000462_190 |
0.123795536 |
- |
- |
PREDICTED: thaumatin-like protein 1 [Jatropha curcas] |
| 11 |
Hb_003894_180 |
0.1262138495 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 12 |
Hb_014361_040 |
0.127657942 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 13 |
Hb_026198_020 |
0.1291551781 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 14 |
Hb_000329_010 |
0.1304348727 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 15 |
Hb_004667_030 |
0.1314559125 |
- |
- |
hypothetical protein VITISV_034568 [Vitis vinifera] |
| 16 |
Hb_004893_010 |
0.1331159609 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 17 |
Hb_007370_010 |
0.133195826 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 18 |
Hb_144449_010 |
0.1333352191 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 19 |
Hb_000152_570 |
0.1345526592 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 20 |
Hb_002809_080 |
0.1348675244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |