| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
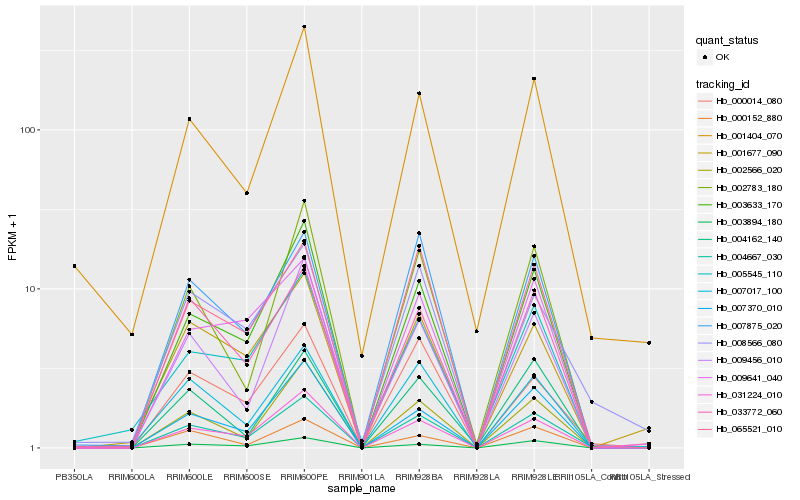
Hb_004667_030 |
0.0 |
- |
- |
hypothetical protein VITISV_034568 [Vitis vinifera] |
| 2 |
Hb_003633_170 |
0.0950078467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_033772_060 |
0.1009888485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_031224_010 |
0.1029203042 |
- |
- |
- |
| 5 |
Hb_065521_010 |
0.1105712313 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |
| 6 |
Hb_002566_020 |
0.1140844837 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 7 |
Hb_001677_090 |
0.1153700281 |
- |
- |
PREDICTED: CASP-like protein 5B2 [Jatropha curcas] |
| 8 |
Hb_007017_100 |
0.1158898691 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007875_020 |
0.1161251512 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 10 |
Hb_009456_010 |
0.1163556184 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X3 [Populus euphratica] |
| 11 |
Hb_003894_180 |
0.1171529427 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 12 |
Hb_007370_010 |
0.1184508527 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 13 |
Hb_002783_180 |
0.1211779542 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |
| 14 |
Hb_004162_140 |
0.121659671 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 15 |
Hb_008566_080 |
0.1225255822 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 16 |
Hb_000014_080 |
0.1261174201 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 17 |
Hb_001404_070 |
0.1296922807 |
- |
- |
sucrose synthase 4 [Hevea brasiliensis] |
| 18 |
Hb_005545_110 |
0.1314559125 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 19 |
Hb_009641_040 |
0.1321959968 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 20 |
Hb_000152_880 |
0.1331590825 |
- |
- |
- |