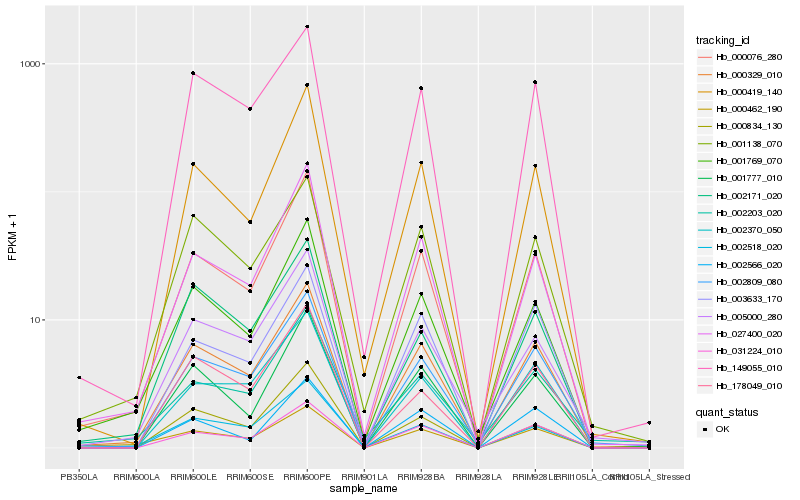
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 2 |
Hb_002809_080 |
0.054458695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000076_280 |
0.0661337361 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 4 |
Hb_001769_070 |
0.0749914449 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_031224_010 |
0.0797935133 |
- |
- |
- |
| 6 |
Hb_149055_010 |
0.0839868521 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 7 |
Hb_000419_140 |
0.0840583052 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 8 |
Hb_027400_020 |
0.0878475333 |
- |
- |
PREDICTED: uncharacterized protein At5g22580 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002370_050 |
0.094085172 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 10 |
Hb_001138_070 |
0.0942415106 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_001777_010 |
0.0954090725 |
- |
- |
- |
| 12 |
Hb_005000_280 |
0.0966218023 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 13 |
Hb_002518_020 |
0.1010294548 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 14 |
Hb_003633_170 |
0.1039475587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_178049_010 |
0.1135258277 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000462_190 |
0.1159055702 |
- |
- |
PREDICTED: thaumatin-like protein 1 [Jatropha curcas] |
| 17 |
Hb_002171_020 |
0.1162036002 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 18 |
Hb_002566_020 |
0.118682575 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 19 |
Hb_002203_020 |
0.1188008233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000834_130 |
0.1200372877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |