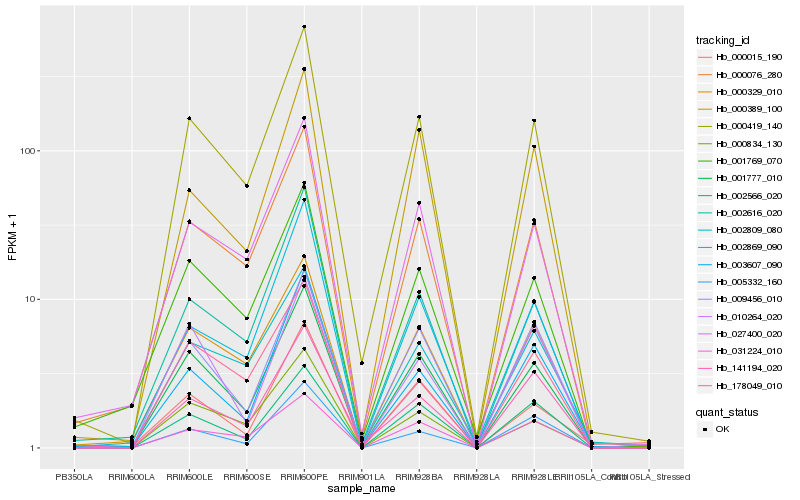
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001777_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000419_140 |
0.0609740253 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 3 |
Hb_000076_280 |
0.0906906147 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 4 |
Hb_002566_020 |
0.0938997592 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 5 |
Hb_000329_010 |
0.0954090725 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 6 |
Hb_009456_010 |
0.0972197921 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X3 [Populus euphratica] |
| 7 |
Hb_001769_070 |
0.1004740882 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000834_130 |
0.1012502832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_027400_020 |
0.1014978697 |
- |
- |
PREDICTED: uncharacterized protein At5g22580 isoform X2 [Jatropha curcas] |
| 10 |
Hb_031224_010 |
0.1084671329 |
- |
- |
- |
| 11 |
Hb_002616_020 |
0.1193119573 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 12 |
Hb_000389_100 |
0.1201486791 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 13 |
Hb_010264_020 |
0.1222803697 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_002809_080 |
0.1231384066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000015_190 |
0.1269294415 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |
| 16 |
Hb_003607_090 |
0.1277625095 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 17 |
Hb_178049_010 |
0.1279659813 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002869_090 |
0.1280880636 |
- |
- |
PREDICTED: dirigent protein 22-like [Jatropha curcas] |
| 19 |
Hb_005332_160 |
0.131790309 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 20 |
Hb_141194_020 |
0.1324339932 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |