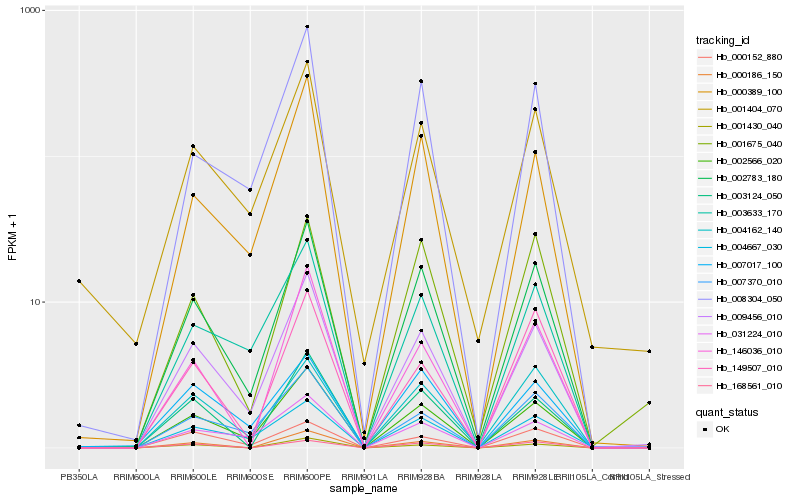
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_180 |
0.0 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |
| 2 |
Hb_002566_020 |
0.0576258671 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 3 |
Hb_009456_010 |
0.0701179297 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X3 [Populus euphratica] |
| 4 |
Hb_008304_050 |
0.1013892692 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 5 |
Hb_004162_140 |
0.1020295637 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 6 |
Hb_000389_100 |
0.1056982822 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 7 |
Hb_000186_150 |
0.1097904366 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 8 |
Hb_003633_170 |
0.1116807991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007370_010 |
0.1127932228 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 10 |
Hb_001675_040 |
0.1155739387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_031224_010 |
0.1195726401 |
- |
- |
- |
| 12 |
Hb_003124_050 |
0.1209309849 |
- |
- |
PREDICTED: uncharacterized protein LOC105641435 [Jatropha curcas] |
| 13 |
Hb_004667_030 |
0.1211779542 |
- |
- |
hypothetical protein VITISV_034568 [Vitis vinifera] |
| 14 |
Hb_007017_100 |
0.1266934469 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001430_040 |
0.1332543626 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 16 |
Hb_001404_070 |
0.1336210989 |
- |
- |
sucrose synthase 4 [Hevea brasiliensis] |
| 17 |
Hb_146036_010 |
0.136164091 |
- |
- |
hypothetical protein B456_005G052500 [Gossypium raimondii] |
| 18 |
Hb_000152_880 |
0.1366091014 |
- |
- |
- |
| 19 |
Hb_168561_010 |
0.1383889003 |
- |
- |
PREDICTED: notchless protein homolog [Jatropha curcas] |
| 20 |
Hb_149507_010 |
0.1390193152 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Malus domestica] |