| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
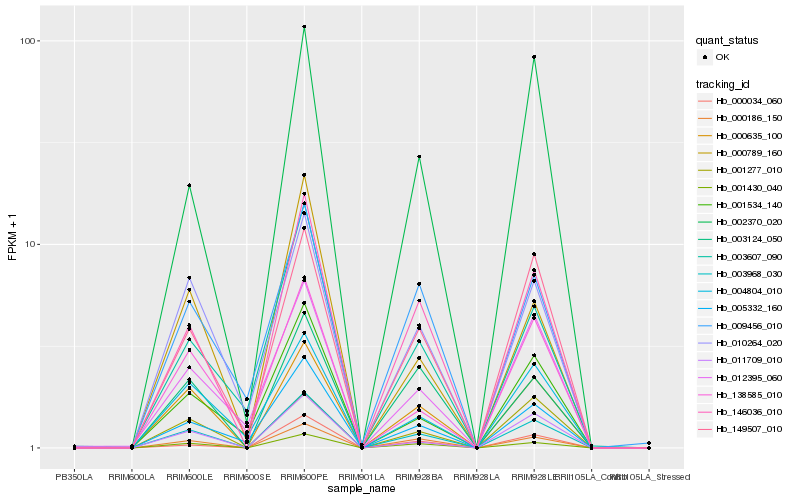
| 1 |
Hb_003968_030 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_001430_040 |
0.0656617128 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 3 |
Hb_146036_010 |
0.0677190932 |
- |
- |
hypothetical protein B456_005G052500 [Gossypium raimondii] |
| 4 |
Hb_000635_100 |
0.0750012209 |
- |
- |
- |
| 5 |
Hb_000186_150 |
0.0799601273 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 6 |
Hb_149507_010 |
0.1107990793 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Malus domestica] |
| 7 |
Hb_005332_160 |
0.1129565802 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 8 |
Hb_011709_010 |
0.1166661196 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 9 |
Hb_010264_020 |
0.1229485626 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_003124_050 |
0.126475346 |
- |
- |
PREDICTED: uncharacterized protein LOC105641435 [Jatropha curcas] |
| 11 |
Hb_002370_020 |
0.1288521368 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 12 |
Hb_001534_140 |
0.1314278413 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 13 |
Hb_004804_010 |
0.13645227 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 14 |
Hb_012395_060 |
0.1382838817 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 15 |
Hb_000034_060 |
0.1404926127 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 16 |
Hb_001277_010 |
0.140778347 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_003607_090 |
0.1419752992 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 18 |
Hb_000789_160 |
0.142765229 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: leucine-rich repeat extensin-like protein 4 [Jatropha curcas] |
| 19 |
Hb_138585_010 |
0.1446904594 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 20 |
Hb_009456_010 |
0.1458693334 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X3 [Populus euphratica] |