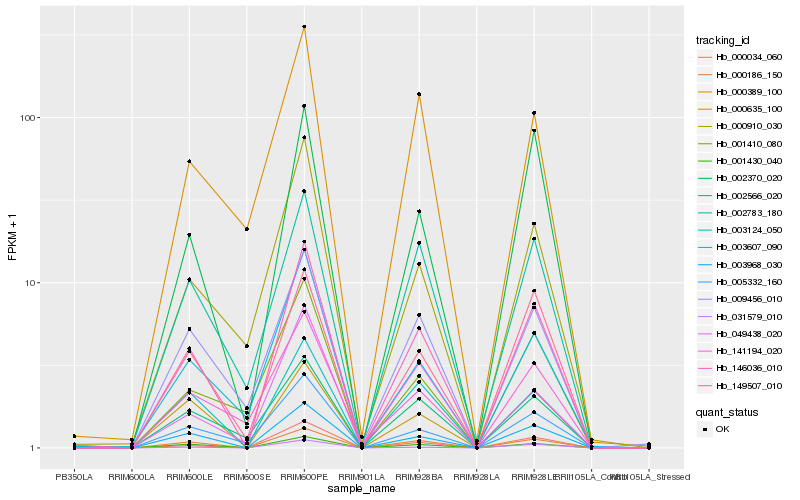
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146036_010 |
0.0 |
- |
- |
hypothetical protein B456_005G052500 [Gossypium raimondii] |
| 2 |
Hb_000186_150 |
0.063637171 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 3 |
Hb_003968_030 |
0.0677190932 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_001430_040 |
0.0791223741 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 5 |
Hb_000034_060 |
0.1001783941 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 6 |
Hb_003124_050 |
0.111692034 |
- |
- |
PREDICTED: uncharacterized protein LOC105641435 [Jatropha curcas] |
| 7 |
Hb_005332_160 |
0.1202291022 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 8 |
Hb_000635_100 |
0.1226267171 |
- |
- |
- |
| 9 |
Hb_002370_020 |
0.1253367936 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 10 |
Hb_149507_010 |
0.1255318236 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Malus domestica] |
| 11 |
Hb_002783_180 |
0.136164091 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |
| 12 |
Hb_002566_020 |
0.136296647 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 13 |
Hb_003607_090 |
0.1365909565 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 14 |
Hb_009456_010 |
0.1367696406 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X3 [Populus euphratica] |
| 15 |
Hb_000910_030 |
0.1413435728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_031579_010 |
0.1516557235 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 17 |
Hb_049438_020 |
0.1524684781 |
- |
- |
PREDICTED: peroxidase 20 [Jatropha curcas] |
| 18 |
Hb_000389_100 |
0.1531805716 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 19 |
Hb_001410_080 |
0.153622443 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_141194_020 |
0.1560868395 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |